You are browsing environment: FUNGIDB
CAZyme Information: AKAW_07147-t41_1-p1
Basic Information
help
| Species |
Aspergillus luchuensis
|
| Lineage |
Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus luchuensis
|
| CAZyme ID |
AKAW_07147-t41_1-p1
|
| CAZy Family |
GT71 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 247 |
|
28676.48 |
6.9213 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_AluchuensisIFO4308 |
11755 |
1033177 |
267 |
11488
|
|
| Gene Location |
No EC number prediction in AKAW_07147-t41_1-p1.
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 226297
|
OCH1 |
4.92e-07 |
96 |
175 |
79 |
154 |
Mannosyltransferase OCH1 or related enzyme [Cell wall/membrane/envelope biogenesis]. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 8.29e-130 |
1 |
175 |
1 |
175 |
| 8.29e-130 |
1 |
175 |
1 |
175 |
| 1.41e-110 |
1 |
175 |
1 |
176 |
| 2.04e-61 |
7 |
175 |
6 |
174 |
| 5.73e-54 |
7 |
175 |
6 |
175 |
AKAW_07147-t41_1-p1 has no PDB hit.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 8.17e-06 |
97 |
173 |
55 |
127 |
Inositol phosphoceramide mannosyltransferase 3 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPAC17G8.11c PE=1 SV=1 |
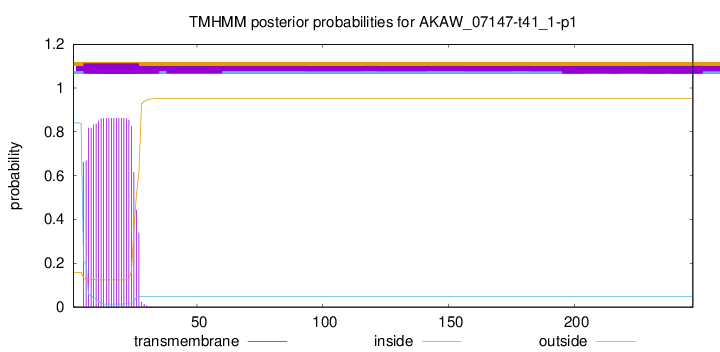
This protein is predicted as OTHER

| Other |
SP_Sec_SPI |
CS Position |
| 0.999742 |
0.000288 |
|