You are browsing environment: FUNGIDB
CAZyme Information: AKAW_05592-t41_1-p1
You are here: Home > Sequence: AKAW_05592-t41_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
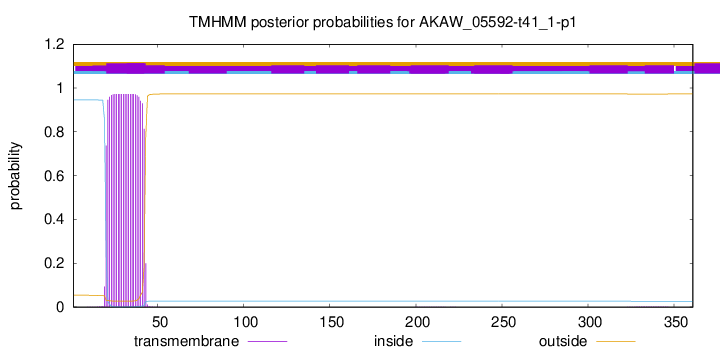
TMHMM annotations
Basic Information help
| Species | Aspergillus luchuensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus luchuensis | |||||||||||
| CAZyme ID | AKAW_05592-t41_1-p1 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE3 | 176 | 330 | 1.8e-34 | 0.7371134020618557 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 238871 | XynB_like | 3.70e-42 | 181 | 333 | 14 | 157 | SGNH_hydrolase subfamily, similar to Ruminococcus flavefaciens XynB. Most likely a secreted hydrolase with xylanase activity. SGNH hydrolases are a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad found in other serine hydrolases. |
| 404371 | Lipase_GDSL_2 | 6.33e-08 | 200 | 325 | 54 | 176 | GDSL-like Lipase/Acylhydrolase family. This family of presumed lipases and related enzymes are similar to pfam00657. |
| 238141 | SGNH_hydrolase | 4.41e-07 | 183 | 329 | 39 | 184 | SGNH_hydrolase, or GDSL_hydrolase, is a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the typical Ser-His-Asp(Glu) triad from other serine hydrolases, but may lack the carboxlic acid. |
| 238872 | SGNH_hydrolase_like_2 | 8.33e-05 | 190 | 330 | 45 | 189 | SGNH_hydrolase subfamily. SGNH hydrolases are a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad found in other serine hydrolases. |
| 238868 | XynE_like | 6.18e-04 | 201 | 333 | 68 | 204 | SGNH_hydrolase subfamily, similar to the putative arylesterase/acylhydrolase from the rumen anaerobe Prevotella bryantii XynE. The P. bryantii XynE gene is located in a xylanase gene cluster. SGNH hydrolases are a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad found in other serine hydrolases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.63e-201 | 1 | 361 | 23 | 395 | |
| 2.95e-93 | 135 | 354 | 89 | 311 | |
| 8.05e-93 | 139 | 354 | 15 | 222 | |
| 1.79e-68 | 142 | 354 | 551 | 769 | |
| 1.89e-58 | 142 | 354 | 375 | 591 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
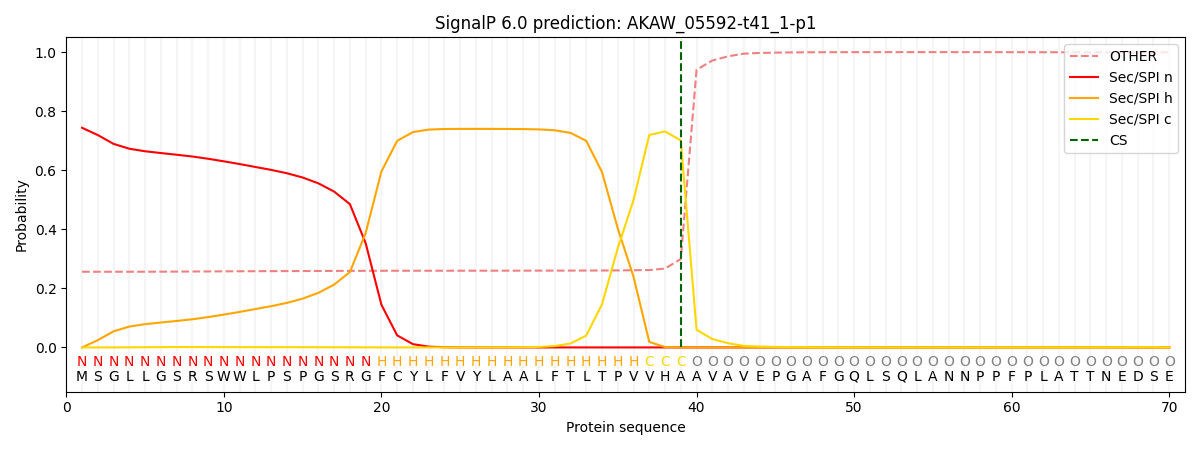
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.273797 | 0.726177 | CS pos: 39-40. Pr: 0.7006 |