You are browsing environment: FUNGIDB
CAZyme Information: AKAW_05253-t41_1-p1
Basic Information
help
| Species |
Aspergillus luchuensis
|
| Lineage |
Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus luchuensis
|
| CAZyme ID |
AKAW_05253-t41_1-p1
|
| CAZy Family |
GH16 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 684 |
DF126458|CGC11 |
76661.00 |
5.0266 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_AluchuensisIFO4308 |
11755 |
1033177 |
267 |
11488
|
|
| Gene Location |
No EC number prediction in AKAW_05253-t41_1-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| PL27 |
62 |
674 |
3.2e-219 |
0.9950819672131147 |
AKAW_05253-t41_1-p1 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 0.0 |
1 |
684 |
1 |
684 |
| 0.0 |
1 |
684 |
1 |
684 |
| 0.0 |
1 |
684 |
1 |
684 |
| 0.0 |
1 |
684 |
1 |
684 |
| 1.15e-195 |
28 |
680 |
20 |
676 |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 4.84e-55 |
66 |
674 |
69 |
688 |
Polysaccharide Lyase BACCELL_00875 [Bacteroides cellulosilyticus DSM 14838],5NO8_B Polysaccharide Lyase BACCELL_00875 [Bacteroides cellulosilyticus DSM 14838] |
| 6.58e-52 |
66 |
674 |
69 |
688 |
Polysaccharide Lyase BACCELL_00875 [Bacteroides cellulosilyticus DSM 14838],5NOK_B Polysaccharide Lyase BACCELL_00875 [Bacteroides cellulosilyticus DSM 14838] |
AKAW_05253-t41_1-p1 has no Swissprot hit.
This protein is predicted as SP
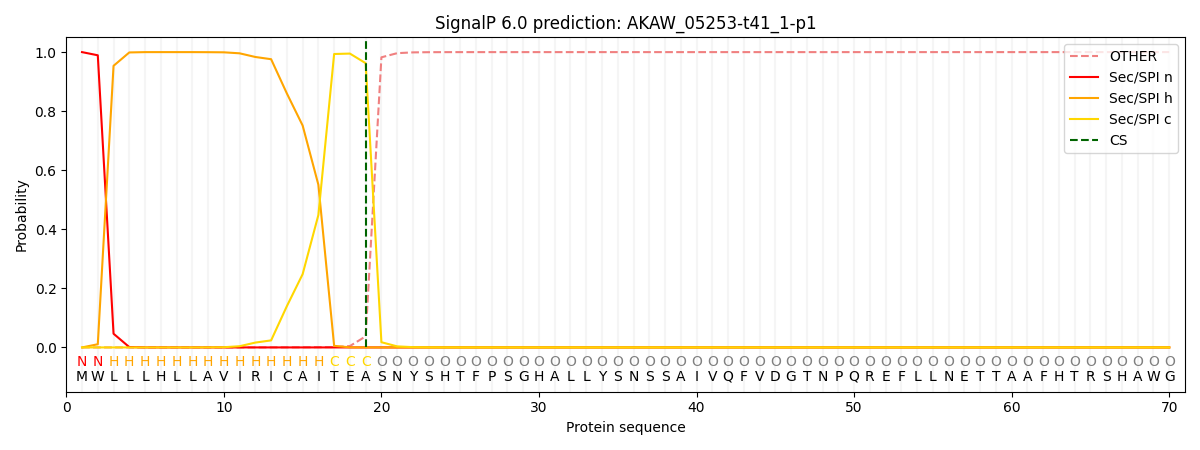
| Other |
SP_Sec_SPI |
CS Position |
| 0.000186 |
0.999773 |
CS pos: 19-20. Pr: 0.9625 |
There is no transmembrane helices in AKAW_05253-t41_1-p1.

