You are browsing environment: FUNGIDB
CAZyme Information: AKAW_05024-t41_1-p1
Basic Information
help
| Species |
Aspergillus luchuensis
|
| Lineage |
Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus luchuensis
|
| CAZyme ID |
AKAW_05024-t41_1-p1
|
| CAZy Family |
GH135 |
| CAZyme Description |
six-hairpin glycosidase
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 679 |
DF126457|CGC6 |
75812.11 |
4.1351 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_AluchuensisIFO4308 |
11755 |
1033177 |
267 |
11488
|
|
| Gene Location |
No EC number prediction in AKAW_05024-t41_1-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| GH142 |
30 |
511 |
3.4e-157 |
0.9624217118997912 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 215307
|
PLN02567 |
0.005 |
216 |
279 |
331 |
384 |
alpha,alpha-trehalase |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 0.0 |
1 |
679 |
1 |
679 |
| 0.0 |
1 |
679 |
1 |
679 |
| 0.0 |
12 |
679 |
13 |
678 |
| 0.0 |
12 |
679 |
30 |
695 |
| 0.0 |
12 |
679 |
13 |
678 |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 4.15e-69 |
34 |
515 |
653 |
1104 |
Sialidase BT_1020 [Bacteroides thetaiotaomicron] |
| 1.69e-65 |
34 |
515 |
653 |
1104 |
Sialidase BT_1020 [Bacteroides thetaiotaomicron] |
| 5.27e-13 |
34 |
512 |
305 |
809 |
Crystal structure of GH121 beta-L-arabinobiosidase HypBA2 from Bifidobacterium longum [Bifidobacterium longum] |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 4.16e-16 |
34 |
679 |
336 |
1048 |
Beta-L-arabinobiosidase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=hypBA2 PE=1 SV=1 |
This protein is predicted as SP
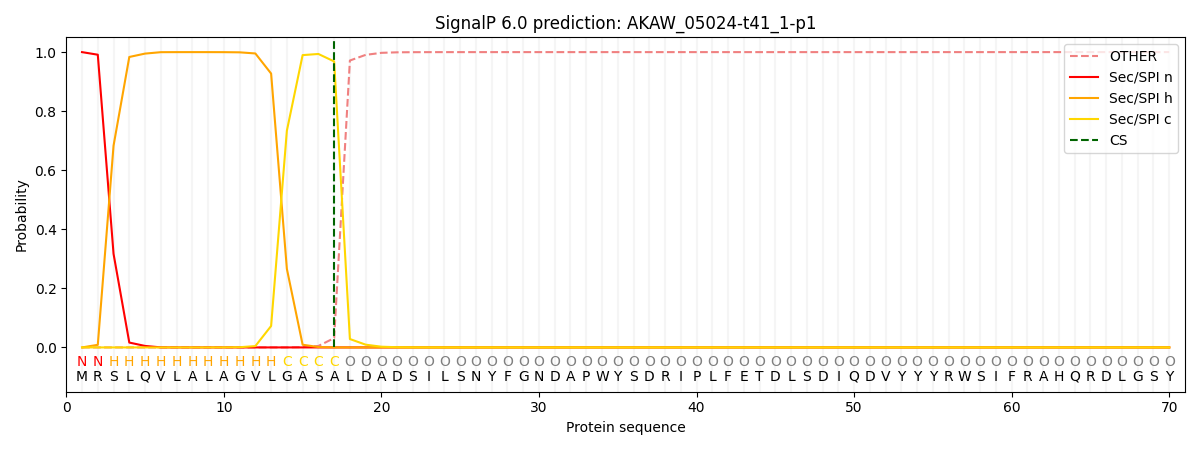
| Other |
SP_Sec_SPI |
CS Position |
| 0.000237 |
0.999720 |
CS pos: 17-18. Pr: 0.9684 |
There is no transmembrane helices in AKAW_05024-t41_1-p1.

