You are browsing environment: FUNGIDB
CAZyme Information: AKAW_02571-t41_1-p1
Basic Information
help
| Species |
Aspergillus luchuensis
|
| Lineage |
Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus luchuensis
|
| CAZyme ID |
AKAW_02571-t41_1-p1
|
| CAZy Family |
AA7 |
| CAZyme Description |
extracellular serine-rich protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 804 |
|
86048.12 |
4.4100 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_AluchuensisIFO4308 |
11755 |
1033177 |
267 |
11488
|
|
| Gene Location |
| Family |
Start |
End |
Evalue |
family coverage |
| CE18 |
367 |
734 |
5.1e-189 |
0.9945504087193461 |
| CBM87 |
149 |
366 |
1.7e-111 |
0.9954128440366973 |
AKAW_02571-t41_1-p1 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 0.0 |
1 |
804 |
1 |
804 |
| 0.0 |
1 |
804 |
1 |
804 |
| 0.0 |
1 |
804 |
1 |
804 |
| 0.0 |
140 |
804 |
134 |
794 |
| 0.0 |
140 |
804 |
25 |
685 |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 0.0 |
140 |
804 |
21 |
684 |
Crystal structure of Agd3 a novel carbohydrate deacetylase [Aspergillus fumigatus Af293] |
AKAW_02571-t41_1-p1 has no Swissprot hit.
This protein is predicted as OTHER
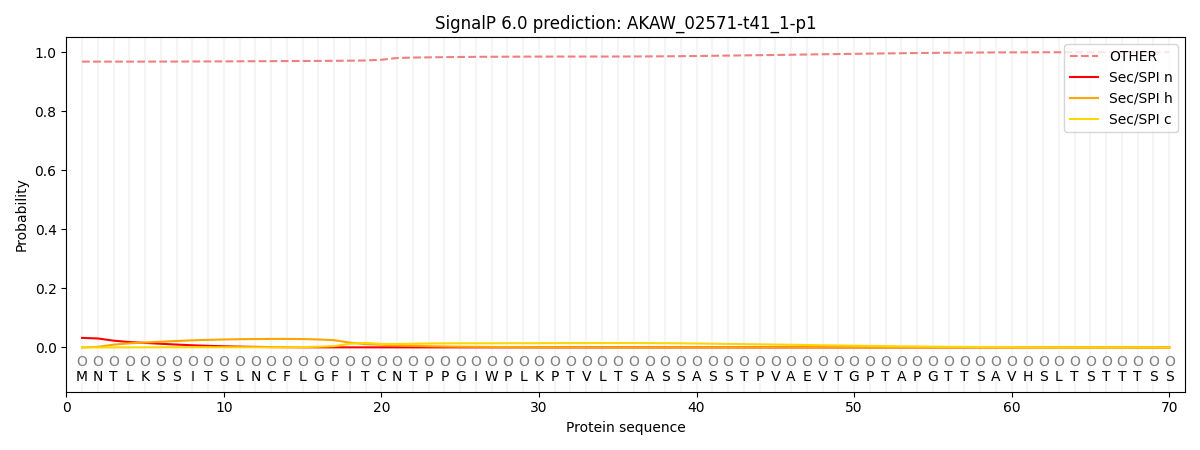
| Other |
SP_Sec_SPI |
CS Position |
| 0.969277 |
0.030731 |
|
There is no transmembrane helices in AKAW_02571-t41_1-p1.

