You are browsing environment: FUNGIDB
CAZyme Information: AKAW_01212-t41_1-p1
Basic Information
help
| Species |
Aspergillus luchuensis
|
| Lineage |
Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus luchuensis
|
| CAZyme ID |
AKAW_01212-t41_1-p1
|
| CAZy Family |
AA3 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 998 |
DF126448|CGC6 |
109113.66 |
4.5273 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_AluchuensisIFO4308 |
11755 |
1033177 |
267 |
11488
|
|
| Gene Location |
No EC number prediction in AKAW_01212-t41_1-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| GH106 |
256 |
766 |
1.5e-55 |
0.6189320388349514 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 407266
|
Glyco_hydro_106 |
1.05e-12 |
259 |
735 |
401 |
869 |
alpha-L-rhamnosidase. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 0.0 |
1 |
998 |
1 |
998 |
| 0.0 |
1 |
998 |
1 |
998 |
| 0.0 |
8 |
995 |
10 |
999 |
| 0.0 |
28 |
798 |
53 |
805 |
| 0.0 |
22 |
994 |
18 |
1018 |
AKAW_01212-t41_1-p1 has no PDB hit.
AKAW_01212-t41_1-p1 has no Swissprot hit.
This protein is predicted as SP
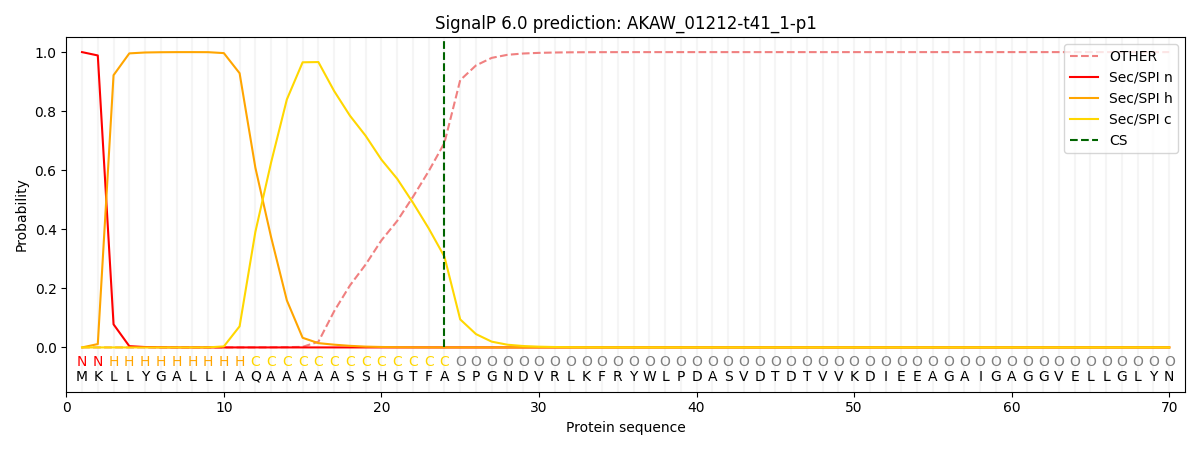
| Other |
SP_Sec_SPI |
CS Position |
| 0.000448 |
0.999519 |
CS pos: 24-25. Pr: 0.3073 |
There is no transmembrane helices in AKAW_01212-t41_1-p1.

