You are browsing environment: FUNGIDB
CAZyme Information: AGR95_077020.mRNA-p1
You are here: Home > Sequence: AGR95_077020.mRNA-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Histoplasma capsulatum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Ajellomycetaceae; Histoplasma; Histoplasma capsulatum | |||||||||||
| CAZyme ID | AGR95_077020.mRNA-p1 | |||||||||||
| CAZy Family | GT1 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.-:14 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT50 | 136 | 410 | 7.3e-92 | 0.9923664122137404 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 178434 | PLN02841 | 1.48e-87 | 7 | 411 | 8 | 415 | GPI mannosyltransferase |
| 252941 | Mannosyl_trans | 1.87e-71 | 136 | 411 | 1 | 259 | Mannosyltransferase (PIG-M). PIG-M has a DXD motif. The DXD motif is found in many glycosyltransferases that utilize nucleotide sugars. It is thought that the motif is involved in the binding of a manganese ion that is required for association of the enzymes with nucleotide sugar substrates. |
| 399599 | PIG-U | 1.15e-04 | 55 | 256 | 47 | 243 | GPI transamidase subunit PIG-U. Many eukaryotic proteins are anchored to the cell surface via glycosylphosphatidylinositol (GPI), which is posttranslationally attached to the carboxyl-terminus by GPI transamidase. The mammalian GPI transamidase is a complex of at least four subunits, GPI8, GAA1, PIG-S, and PIG-T. PIG-U is thought to represent a fifth subunit in this complex and may be involved in the recognition of either the GPI attachment signal or the lipid portion of GPI. |
| 401508 | GT87 | 0.008 | 66 | 179 | 3 | 113 | Glycosyltransferase family 87. The enzymes in this family are glycosyltransferases. PimE is involved in phosphatidylinositol mannoside (PIM) synthesis, a major class of glycolipids in all mycobacteria. PimE is a polyprenol-phosphate-mannose-dependent mannosyltransferase that transfers the fifth mannose of PIM. The family also includes alpha(1-->3) arabinofuranosyltransferase, invloved in the synthesis of of mycobacterial arabinogalactan. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.52e-291 | 1 | 424 | 1 | 429 | |
| 3.41e-289 | 1 | 424 | 1 | 429 | |
| 5.65e-288 | 1 | 424 | 1 | 429 | |
| 9.34e-181 | 1 | 414 | 1 | 415 | |
| 7.33e-179 | 1 | 418 | 1 | 414 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.30e-179 | 1 | 418 | 1 | 414 | GPI mannosyltransferase 1 OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=gpi14 PE=3 SV=2 |
|
| 1.08e-173 | 1 | 414 | 1 | 421 | GPI mannosyltransferase 1 OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=gpi14 PE=3 SV=1 |
|
| 4.71e-172 | 1 | 414 | 1 | 428 | GPI mannosyltransferase 1 OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=gpi14 PE=3 SV=1 |
|
| 3.77e-161 | 1 | 421 | 1 | 415 | GPI mannosyltransferase 1 OS=Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1) OX=229533 GN=GPI14 PE=3 SV=1 |
|
| 3.21e-158 | 1 | 413 | 19 | 484 | GPI mannosyltransferase 1 OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=gim-1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000023 | 0.000001 |
TMHMM Annotations download full data without filtering help
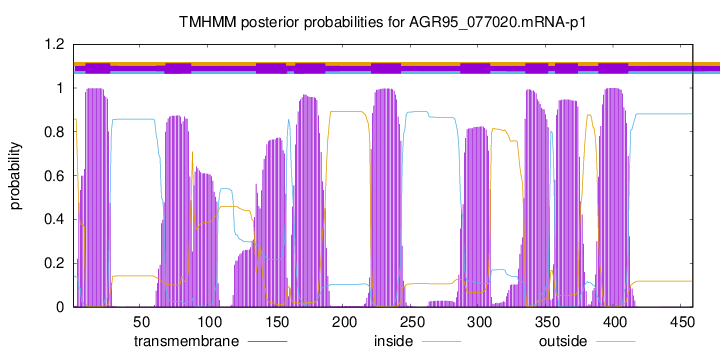
| Start | End |
|---|---|
| 10 | 28 |
| 69 | 88 |
| 136 | 158 |
| 165 | 187 |
| 221 | 243 |
| 287 | 309 |
| 335 | 352 |
| 357 | 374 |
| 389 | 411 |
