You are browsing environment: FUNGIDB
CAZyme Information: AGR95_021250.mRNA-p1
You are here: Home > Sequence: AGR95_021250.mRNA-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
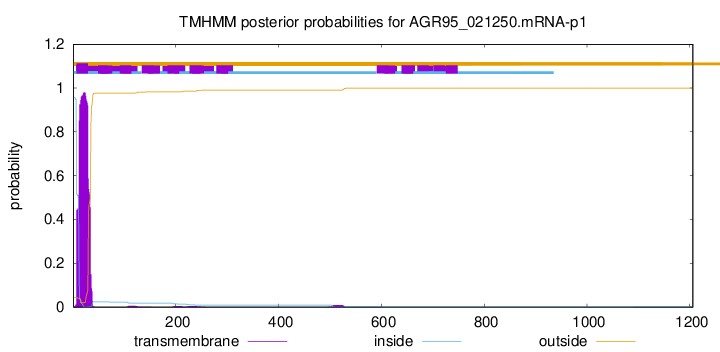
TMHMM annotations
Basic Information help
| Species | Histoplasma capsulatum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Ajellomycetaceae; Histoplasma; Histoplasma capsulatum | |||||||||||
| CAZyme ID | AGR95_021250.mRNA-p1 | |||||||||||
| CAZy Family | AA7 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.186:3 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT8 | 533 | 664 | 4.2e-22 | 0.5758754863813229 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 133018 | GT8_Glycogenin | 6.20e-41 | 534 | 664 | 6 | 133 | Glycogenin belongs the GT 8 family and initiates the biosynthesis of glycogen. Glycogenin initiates the biosynthesis of glycogen by incorporating glucose residues through a self-glucosylation reaction at a Tyr residue, and then acts as substrate for chain elongation by glycogen synthase and branching enzyme. It contains a conserved DxD motif and an N-terminal beta-alpha-beta Rossmann-like fold that are common to the nucleotide-binding domains of most glycosyltransferases. The DxD motif is essential for coordination of the catalytic divalent cation, most commonly Mn2+. Glycogenin can be classified as a retaining glycosyltransferase, based on the relative anomeric stereochemistry of the substrate and product in the reaction catalyzed. It is placed in glycosyltransferase family 8 which includes lipopolysaccharide glucose and galactose transferases and galactinol synthases. |
| 223669 | MhpC | 1.31e-17 | 69 | 400 | 2 | 272 | Pimeloyl-ACP methyl ester carboxylesterase [Coenzyme transport and metabolism, General function prediction only]. |
| 395444 | Abhydrolase_1 | 4.01e-17 | 106 | 394 | 1 | 243 | alpha/beta hydrolase fold. This catalytic domain is found in a very wide range of enzymes. |
| 235628 | PRK05855 | 3.79e-10 | 99 | 222 | 19 | 121 | SDR family oxidoreductase. |
| 235135 | PRK03592 | 1.12e-08 | 108 | 217 | 30 | 116 | haloalkane dehalogenase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 545 | 1158 | 1 | 691 | |
| 0.0 | 538 | 1158 | 16 | 713 | |
| 7.63e-205 | 52 | 1158 | 591 | 1647 | |
| 3.55e-192 | 4 | 664 | 2 | 736 | |
| 1.68e-145 | 538 | 1176 | 15 | 748 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.12e-21 | 535 | 684 | 11 | 151 | Crystal Structure of Human Glycogenin-1 (GYG1) Tyr195pIPhe mutant, apo form [Homo sapiens],6EQL_A Crystal Structure of Human Glycogenin-1 (GYG1) Tyr195pIPhe mutant complexed with manganese and UDP [Homo sapiens],6EQL_B Crystal Structure of Human Glycogenin-1 (GYG1) Tyr195pIPhe mutant complexed with manganese and UDP [Homo sapiens] |
|
| 2.12e-21 | 535 | 684 | 11 | 151 | Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese and UDP, in a triclinic closed form [Homo sapiens],3T7M_B Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese and UDP, in a triclinic closed form [Homo sapiens],3T7N_A Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese and UDP, in a monoclinic closed form [Homo sapiens],3T7N_B Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese and UDP, in a monoclinic closed form [Homo sapiens],3T7O_A Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese, UDP-Glucose and glucose [Homo sapiens],3T7O_B Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese, UDP-Glucose and glucose [Homo sapiens],3U2U_A Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese, UDP and maltotetraose [Homo sapiens],3U2U_B Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese, UDP and maltotetraose [Homo sapiens],3U2V_A Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese, UDP and maltohexaose [Homo sapiens],3U2V_B Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese, UDP and maltohexaose [Homo sapiens],3U2X_A Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese, UDP and 1'-deoxyglucose [Homo sapiens],3U2X_B Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese, UDP and 1'-deoxyglucose [Homo sapiens] |
|
| 2.12e-21 | 535 | 684 | 11 | 151 | Crystal Structure of Human Glycogenin-1 (GYG1), apo form [Homo sapiens],3QVB_A Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese and UDP [Homo sapiens],3U2W_A Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese and glucose or a glucal species [Homo sapiens],3U2W_B Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese and glucose or a glucal species [Homo sapiens] |
|
| 3.23e-21 | 535 | 684 | 32 | 172 | Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese [Homo sapiens] |
|
| 8.95e-21 | 535 | 684 | 30 | 170 | structure of glycogenin truncated at residue 270 in a complex with UDP [Oryctolagus cuniculus],1ZCT_B structure of glycogenin truncated at residue 270 in a complex with UDP [Oryctolagus cuniculus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.87e-24 | 528 | 663 | 3 | 135 | Glycogenin-1 OS=Caenorhabditis elegans OX=6239 GN=gyg-1 PE=1 SV=1 |
|
| 6.56e-21 | 535 | 684 | 10 | 150 | Glycogenin-1 OS=Mus musculus OX=10090 GN=Gyg1 PE=1 SV=3 |
|
| 5.09e-20 | 535 | 684 | 10 | 150 | Glycogenin-1 OS=Homo sapiens OX=9606 GN=GYG1 PE=1 SV=4 |
|
| 5.29e-20 | 535 | 684 | 10 | 150 | Glycogenin-1 OS=Rattus norvegicus OX=10116 GN=Gyg1 PE=2 SV=4 |
|
| 9.60e-20 | 535 | 684 | 10 | 150 | Glycogenin-1 OS=Oryctolagus cuniculus OX=9986 GN=GYG1 PE=1 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000055 | 0.000010 |