You are browsing environment: FUNGIDB
CAZyme Information: AGR57_9286T0-p1
Basic Information
help
| Species |
Phanerochaete chrysosporium
|
| Lineage |
Basidiomycota; Agaricomycetes; ; Phanerochaetaceae; Phanerochaete; Phanerochaete chrysosporium
|
| CAZyme ID |
AGR57_9286T0-p1
|
| CAZy Family |
GT4 |
| CAZyme Description |
Glycoside Hydrolase Family 30 protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 509 |
|
53333.14 |
6.7501 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_PchrysosporiumRP-78 |
13602 |
273507 |
0 |
13602
|
|
| Gene Location |
No EC number prediction in AGR57_9286T0-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| GH30 |
25 |
508 |
1.1e-49 |
0.9424460431654677 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 405300
|
Glyco_hydr_30_2 |
3.08e-27 |
28 |
256 |
13 |
231 |
O-Glycosyl hydrolase family 30. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 2.40e-134 |
22 |
425 |
23 |
413 |
| 5.04e-122 |
22 |
509 |
21 |
484 |
| 2.01e-121 |
22 |
509 |
21 |
484 |
| 2.01e-121 |
22 |
509 |
21 |
484 |
| 1.93e-120 |
22 |
509 |
29 |
483 |
AGR57_9286T0-p1 has no PDB hit.
AGR57_9286T0-p1 has no Swissprot hit.
This protein is predicted as SP
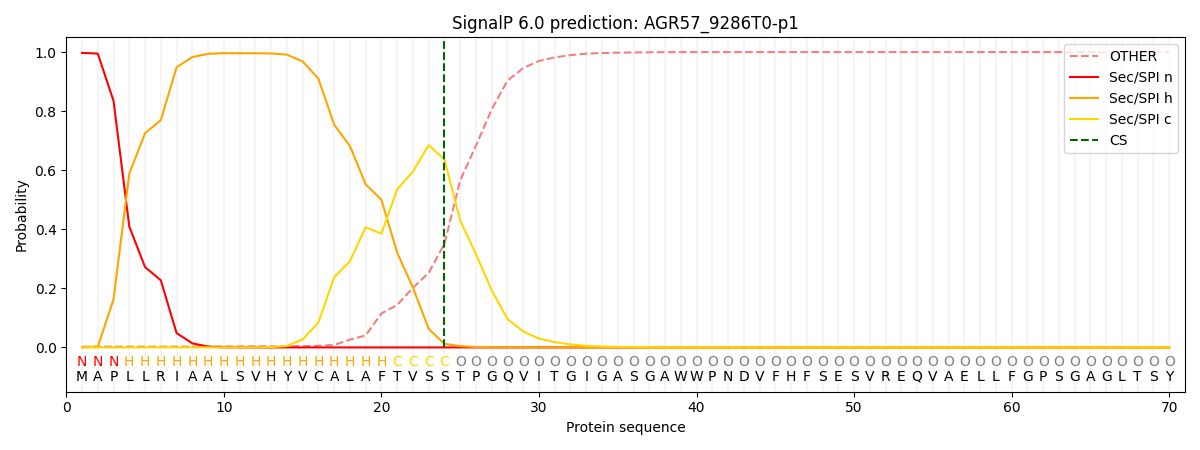
| Other |
SP_Sec_SPI |
CS Position |
| 0.005079 |
0.994912 |
CS pos: 24-25. Pr: 0.6366 |
There is no transmembrane helices in AGR57_9286T0-p1.

