You are browsing environment: FUNGIDB
CAZyme Information: AGR57_9122T0-p1
You are here: Home > Sequence: AGR57_9122T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
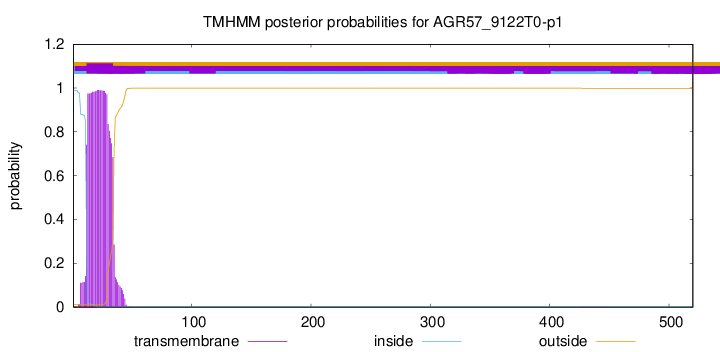
TMHMM annotations
Basic Information help
| Species | Phanerochaete chrysosporium | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Agaricomycetes; ; Phanerochaetaceae; Phanerochaete; Phanerochaete chrysosporium | |||||||||||
| CAZyme ID | AGR57_9122T0-p1 | |||||||||||
| CAZy Family | GT39 | |||||||||||
| CAZyme Description | Glycosyltransferase Family 49 protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT49 | 133 | 488 | 8.2e-36 | 0.8724035608308606 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 404735 | Glyco_transf_49 | 3.46e-66 | 152 | 481 | 1 | 326 | Glycosyl-transferase for dystroglycan. This glycosyl-transferase brings about the glycosylation of the alpha-dystroglycan subunit. Dystroglycan is an integral member of the skeletal muscular dystrophin glycoprotein complex, which links dystrophin to proteins in the extracellular matrix. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.48e-114 | 86 | 513 | 161 | 546 | |
| 1.71e-100 | 1 | 511 | 5 | 429 | |
| 9.92e-100 | 79 | 508 | 49 | 416 | |
| 5.21e-98 | 120 | 513 | 83 | 429 | |
| 9.32e-95 | 118 | 512 | 84 | 423 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.58e-16 | 148 | 481 | 90 | 408 | Beta-1,4-glucuronyltransferase 1 OS=Homo sapiens OX=9606 GN=B4GAT1 PE=1 SV=1 |
|
| 5.58e-16 | 148 | 481 | 90 | 408 | Beta-1,4-glucuronyltransferase 1 OS=Pongo abelii OX=9601 GN=B4GAT1 PE=2 SV=1 |
|
| 9.96e-16 | 148 | 481 | 90 | 408 | Beta-1,4-glucuronyltransferase 1 OS=Bos taurus OX=9913 GN=B4GAT1 PE=2 SV=2 |
|
| 7.53e-15 | 148 | 481 | 90 | 408 | Beta-1,4-glucuronyltransferase 1 OS=Mus musculus OX=10090 GN=B4gat1 PE=1 SV=1 |
|
| 2.05e-14 | 150 | 452 | 399 | 638 | Xylosyl- and glucuronyltransferase LARGE2 OS=Rattus norvegicus OX=10116 GN=Large2 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
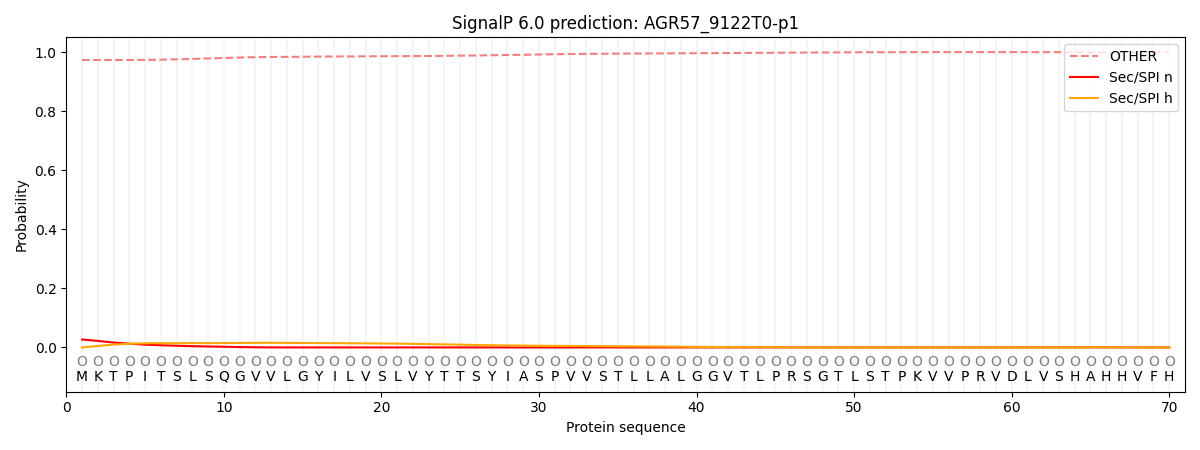
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.974970 | 0.025031 |