You are browsing environment: FUNGIDB
CAZyme Information: AGR57_8978T0-p1
You are here: Home > Sequence: AGR57_8978T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phanerochaete chrysosporium | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Agaricomycetes; ; Phanerochaetaceae; Phanerochaete; Phanerochaete chrysosporium | |||||||||||
| CAZyme ID | AGR57_8978T0-p1 | |||||||||||
| CAZy Family | GT2|GT2 | |||||||||||
| CAZyme Description | Multicopper oxidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA1 | 63 | 530 | 1.9e-92 | 0.9776536312849162 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 274556 | laccase | 8.21e-70 | 46 | 535 | 3 | 518 | laccase, plant. Members of this protein family include the copper-containing enzyme laccase (EC 1.10.3.2), often several from a single plant species, and additional, uncharacterized, closely related plant proteins termed laccase-like multicopper oxidases. This protein family shows considerable sequence similarity to the L-ascorbate oxidase (EC 1.10.3.3) family. Laccases are enzymes of rather broad specificity, and classification of all proteins scoring about the trusted cutoff of this model as laccases may be appropriate. |
| 274555 | ascorbase | 3.83e-69 | 46 | 530 | 1 | 517 | L-ascorbate oxidase, plant type. Members of this protein family are the copper-containing enzyme L-ascorbate oxidase (EC 1.10.3.3), also called ascorbase. This family is found in flowering plants, and shows greater sequence similarity to a family of laccases (EC 1.10.3.2) from plants than to other known ascorbate oxidases. |
| 259953 | CuRO_2_MCO_like_1 | 1.95e-63 | 187 | 341 | 1 | 163 | The second cupredoxin domain of uncharacterized multicopper oxidase. Multicopper Oxidases (MCOs) are multi-domain enzymes that are able to couple oxidation of substrates with reduction of dioxygen to water. MCOs oxidise their substrate by accepting electrons at a mononuclear copper centre and transferring them to a trinuclear copper centre which binds a dioxygen. The dioxygen, following the transfer of four electrons, is reduced to two molecules of water. These MCOs are capable of oxidizing a vast range of substrates, varying from aromatic to inorganic compounds such as metals. This family of MCOs is composed of three cupredoxin domains that include one mononuclear and one trinuclear copper center. The copper ions are bound in several sites: Type 1, Type 2, and/or Type 3. The ensemble of types 2 and 3 copper is called a trinuclear cluster. MCOs oxidize their substrate by accepting electrons at a mononuclear copper center and transferring them to the active site trinuclear copper center. The cupredoxin domain 2 of 3-domain MCOs has lost the ability to bind copper. |
| 259926 | CuRO_1_Diphenol_Ox | 3.06e-63 | 47 | 171 | 1 | 119 | The first cupredoxin domain of fungal laccase, diphenol oxidase. Diphenol oxidase belongs to the laccase family. It catalyzes the initial steps in melanin biosynthesis from diphenols. Melanin is one of the virulence factors of infectious fungi. In the pathogenesis of C. neoformans, melanin pigments have been shown to protect the fungal cells from oxidative and microbicidal activities of host defense systems. Laccase is a blue multicopper oxidase (MCO) which catalyzes the oxidation of a variety aromatic - notably phenolic and inorganic substances coupled to the reduction of molecular oxygen to water. It has been implicated in a wide spectrum of biological activities and, in particular, plays a key role in morphogenesis, development and lignin metabolism. Although MCOs have diverse functions, majority of them have three cupredoxin domain repeats that include one mononuclear and one trinuclear copper center. The copper ions are bound in several sites: Type 1, Type 2, and/or Type 3. The ensemble of types 2 and 3 copper is called a trinuclear cluster. MCOs oxidize their substrate by accepting electrons at a mononuclear copper center and transferring them to the active site trinuclear copper center. The cupredoxin domain 1 of 3-domain MCOs contains part the trinuclear copper binding site, which is located at the interface of domains 1 and 3. |
| 259977 | CuRO_3_MCO_like_4 | 2.70e-56 | 386 | 536 | 2 | 166 | The third cupredoxin domain of uncharacterized multicopper oxidase. Multicopper Oxidases (MCOs) are multi-domain enzymes that are able to couple oxidation of substrates with reduction of dioxygen to water. MCOs oxidize their substrate by accepting electrons at a mononuclear copper centre and transferring them to a trinuclear copper centre which binds a dioxygen. The dioxygen, following the transfer of four electrons, is reduced to two molecules of water. These MCOs are capable of oxidizing a vast range of substrates, varying from aromatic to inorganic compounds such as metals. This subfamily of MCOs is composed of three cupredoxin domains. The cupredoxin domain 3 of 3-domain MCOs contains the Type 1 (T1) copper binding site and part the trinuclear copper binding site, which is located at the interface of domains 1 and 3. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.51e-157 | 13 | 554 | 69 | 614 | |
| 4.44e-149 | 35 | 557 | 104 | 621 | |
| 3.02e-146 | 24 | 554 | 76 | 604 | |
| 1.15e-119 | 43 | 557 | 108 | 661 | |
| 1.11e-115 | 47 | 554 | 123 | 655 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.88e-64 | 60 | 554 | 17 | 491 | Crystal structure of LacB from Trametes sp. AH28-2 [Trametes sp. AH28-2],3KW7_B Crystal structure of LacB from Trametes sp. AH28-2 [Trametes sp. AH28-2] |
|
| 1.39e-63 | 59 | 556 | 17 | 490 | Single crystal serial study of the X-ray induced enzymatic reduction of molecular oxygen to water for laccase from Steccherinum murashkinskyi at sub-atomic resolution. Third structure of the series with 315 KGy dose. [Steccherinum murashkinskyi],6RHI_A Single crystal serial study of the X-ray induced enzymatic reduction of molecular oxygen to water for laccase from Steccherinum murashkinskyi at sub-atomic resolution. Ninth structure of the series with 1215 KGy dose. [Steccherinum murashkinskyi],6RHO_A Single crystal serial study of the X-ray induced enzymatic reduction of molecular oxygen to water for laccase from Steccherinum murashkinskyi at sub-atomic resolution. Twentieth structure of the series with 4065 KGy dose. [Steccherinum murashkinskyi],6RHP_A Single crystal serial study of the X-ray induced enzymatic reduction of molecular oxygen to water for laccase from Steccherinum murashkinskyi at sub-atomic resolution. Twenty first structure of the series with 4415 KGy dose (collected after refreezing). [Steccherinum murashkinskyi],6RHR_A Single crystal serial study of the inhibition of laccases from Steccherinum murashkinskyi by chloride anions at sub-atomic resolution. First structure of the series with 15 KGy dose. [Steccherinum murashkinskyi],6RHU_A Single crystal serial study of the inhibition of laccases from Steccherinum murashkinskyi by chloride anions at sub-atomic resolution. Second structure of the series with 165 KGy dose. [Steccherinum murashkinskyi],6RHX_A Single crystal serial study of the inhibition of laccases from Steccherinum murashkinskyi by chloride anions at sub-atomic resolution. Third structure of the series with 315 KGy dose. [Steccherinum murashkinskyi],6RI0_A Single crystal serial study of the inhibition of laccases from Steccherinum murashkinskyi by chloride anions at sub-atomic resolution. Ninth structure of the series with 1215 KGy dose. [Steccherinum murashkinskyi],6RI2_A Single crystal serial study of the inhibition of laccases from Steccherinum murashkinskyi by chloride anions at sub-atomic resolution. Twentieth structure of the series with 4065 KGy dose. [Steccherinum murashkinskyi],6RI4_A Single crystal serial study of the inhibition of laccases from Steccherinum murashkinskyi by fluoride anions at sub-atomic resolution. First structure of the series with 13 KGy dose. [Steccherinum murashkinskyi],6RI6_A Single crystal serial study of the inhibition of laccases from Steccherinum murashkinskyi by fluoride anions at sub-atomic resolution. Second structure of the series with 400 KGy dose. [Steccherinum murashkinskyi],6RI8_A Single crystal serial study of the inhibition of laccases from Steccherinum murashkinskyi by fluoride anions at sub-atomic resolution. Third structure of the series with 800 KGy dose. [Steccherinum murashkinskyi],6RII_A Single crystal serial study of the inhibition of laccases from Steccherinum murashkinskyi by fluoride anions at sub-atomic resolution. Fourth structure of the series with 1200 KGy dose. [Steccherinum murashkinskyi],6RIK_A Single crystal serial study of the inhibition of laccases from Steccherinum murashkinskyi by fluoride anions at sub-atomic resolution. Thirteenth structure of the series with 5200 KGy dose. [Steccherinum murashkinskyi],6RIL_A Single crystal serial study of the inhibition of laccases from Steccherinum murashkinskyi by fluoride anions at sub-atomic resolution. Fourteenth structure of the series with 5600 KGy dose (data was collected after refreezing). [Steccherinum murashkinskyi] |
|
| 1.42e-63 | 59 | 556 | 17 | 490 | Structural study of the X-ray induced enzymatic reduction of molecular oxygen to water for laccase from Steccherinum murashkinskyi. First structure of the series with 3 min total X-ray exposition time. [Steccherinum murashkinskyi],5MHU_A The study of the X-ray induced enzymatic reduction of molecular oxygen to water for laccase from Steccherinum murashkinskyi.The third structure of the series with total exposition time 63 min. [Steccherinum murashkinskyi],5MHV_A The study of the X-ray induced enzymatic reduction of molecular oxygen to water for laccase from Steccherinum murashkinskyi.The fourth structure of the series with total exposition time 93 min. [Steccherinum murashkinskyi] |
|
| 1.42e-63 | 59 | 556 | 17 | 490 | The study of the X-ray induced enzymatic reduction of molecular oxygen to water for laccase from Steccherinum murashkinskyi. Second structure of the series with total exposition time 33 min. [Steccherinum murashkinskyi],6RGH_A Single crystal serial study of the X-ray induced enzymatic reduction of molecular oxygen to water for laccase from Steccherinum murashkinskyi at sub-atomic resolution. First structure of the series with 15 KGy dose. [Steccherinum murashkinskyi],6RGP_A Single crystal serial study of the X-ray induced enzymatic reduction of molecular oxygen to water for laccase from Steccherinum murashkinskyi at sub-atomic resolution. Second structure of the series with 165 KGy dose. [Steccherinum murashkinskyi] |
|
| 2.66e-63 | 59 | 556 | 17 | 490 | Steccherinum murashkinskyi laccase at 0.95 resolution [Steccherinum murashkinskyi] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.24e-70 | 48 | 556 | 24 | 513 | Iron transport multicopper oxidase fetC OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=fetC PE=2 SV=1 |
|
| 1.30e-66 | 44 | 555 | 59 | 579 | Laccase-1 OS=Cryptococcus neoformans var. neoformans serotype D (strain B-3501A) OX=283643 GN=LAC1 PE=1 SV=1 |
|
| 2.52e-66 | 44 | 555 | 59 | 579 | Laccase-1 OS=Cryptococcus neoformans var. grubii serotype A (strain H99 / ATCC 208821 / CBS 10515 / FGSC 9487) OX=235443 GN=LAC1 PE=1 SV=1 |
|
| 3.45e-63 | 34 | 556 | 10 | 516 | Iron transport multicopper oxidase fetC OS=Epichloe festucae (strain E2368) OX=696363 GN=fetC PE=2 SV=1 |
|
| 4.79e-62 | 47 | 557 | 25 | 518 | Iron transport multicopper oxidase FET3 OS=Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1) OX=229533 GN=FET3 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
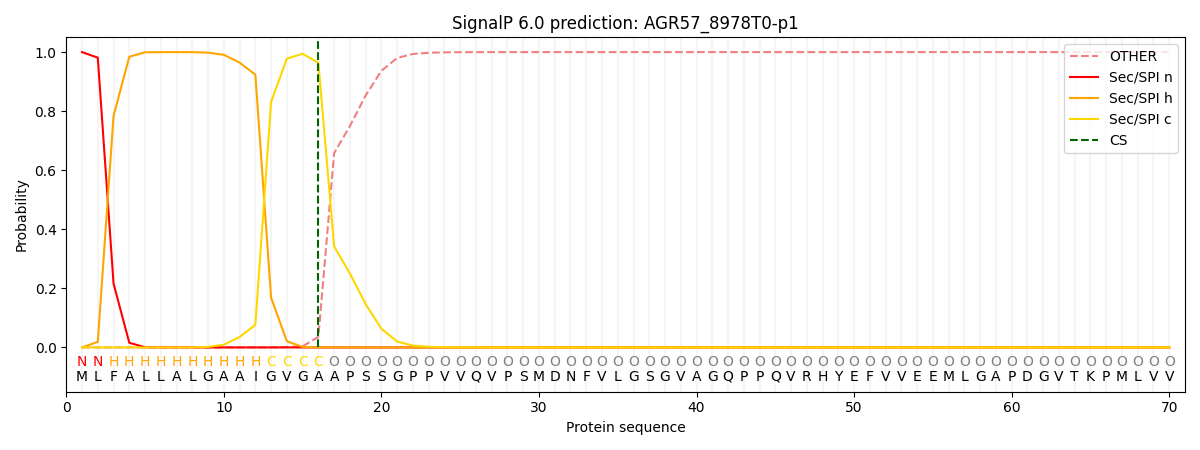
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000298 | 0.999662 | CS pos: 16-17. Pr: 0.9639 |
