You are browsing environment: FUNGIDB
CAZyme Information: AGR57_8506T0-p1
You are here: Home > Sequence: AGR57_8506T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phanerochaete chrysosporium | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Agaricomycetes; ; Phanerochaetaceae; Phanerochaete; Phanerochaete chrysosporium | |||||||||||
| CAZyme ID | AGR57_8506T0-p1 | |||||||||||
| CAZy Family | PL8 | |||||||||||
| CAZyme Description | Carbohydrate-Binding Module Family 1 / Carbohydrate Esterase Family 1 protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.1.1.72:3 |
|---|
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 273828 | esterase_phb | 1.14e-28 | 111 | 292 | 1 | 177 | esterase, PHB depolymerase family. This model describes a subfamily among lipases of the ab-hydrolase family. This subfamily includes bacterial depolymerases for poly(3-hydroxybutyrate) (PHB) and related polyhydroxyalkanoates (PHA), as well as acetyl xylan esterases, feruloyl esterases, and others from fungi. [Fatty acid and phospholipid metabolism, Degradation] |
| 402228 | Esterase_phd | 5.25e-19 | 114 | 241 | 6 | 135 | Esterase PHB depolymerase. This family of proteins include acetyl xylan esterases (AXE), feruloyl esterases (FAE), and poly(3-hydroxybutyrate) (PHB) depolymerases. |
| 226040 | LpqC | 9.51e-16 | 93 | 248 | 31 | 190 | Poly(3-hydroxybutyrate) depolymerase [Secondary metabolites biosynthesis, transport and catabolism]. |
| 197593 | fCBD | 1.03e-09 | 20 | 53 | 1 | 34 | Fungal-type cellulose-binding domain. Small four-cysteine cellulose-binding domain of fungi |
| 395595 | CBM_1 | 1.35e-09 | 22 | 49 | 2 | 29 | Fungal cellulose binding domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.94e-203 | 1 | 293 | 1 | 293 | |
| 1.62e-154 | 1 | 292 | 1 | 290 | |
| 8.65e-150 | 2 | 292 | 4 | 301 | |
| 3.72e-140 | 1 | 292 | 1 | 295 | |
| 2.18e-103 | 10 | 293 | 12 | 280 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.81e-71 | 93 | 293 | 1 | 201 | Acetyl xylan esterase from Aspergillus awamori [Aspergillus awamori],5X6S_B Acetyl xylan esterase from Aspergillus awamori [Aspergillus awamori] |
|
| 7.57e-10 | 17 | 197 | 3 | 207 | Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor covalent complex with the aldouronic acid UXXR [Cerrena unicolor],6RV8_B Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor covalent complex with the aldouronic acid UXXR [Cerrena unicolor] |
|
| 1.86e-08 | 22 | 53 | 5 | 36 | Chain A, Exoglucanase 1 [Trichoderma reesei] |
|
| 1.86e-08 | 22 | 53 | 5 | 36 | Chain A, C-TERMINAL DOMAIN OF CELLOBIOHYDROLASE I [Trichoderma reesei],2CBH_A Chain A, C-TERMINAL DOMAIN OF CELLOBIOHYDROLASE I [Trichoderma reesei],2MWJ_A Chain A, Exoglucanase 1 [Trichoderma reesei],2MWK_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X34_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X35_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X36_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X37_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X38_A Chain A, Exoglucanase 1 [Trichoderma reesei] |
|
| 3.51e-08 | 23 | 53 | 6 | 36 | Chain A, CELLOBIOHYDROLASE I [Trichoderma reesei],1AZH_A Chain A, CELLOBIOHYDROLASE I [Trichoderma reesei],5X3C_A Chain A, Exoglucanase 1 [Trichoderma reesei] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.35e-78 | 91 | 292 | 29 | 230 | Probable acetylxylan esterase A OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=axeA PE=3 SV=1 |
|
| 5.50e-77 | 87 | 293 | 25 | 232 | Probable acetylxylan esterase A OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=axeA PE=3 SV=1 |
|
| 7.09e-77 | 96 | 292 | 32 | 228 | Acetylxylan esterase A OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=axeA PE=1 SV=2 |
|
| 9.36e-77 | 91 | 293 | 29 | 231 | Probable acetylxylan esterase A OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=axeA PE=3 SV=1 |
|
| 4.41e-76 | 87 | 293 | 25 | 232 | Acetylxylan esterase A OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=axeA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
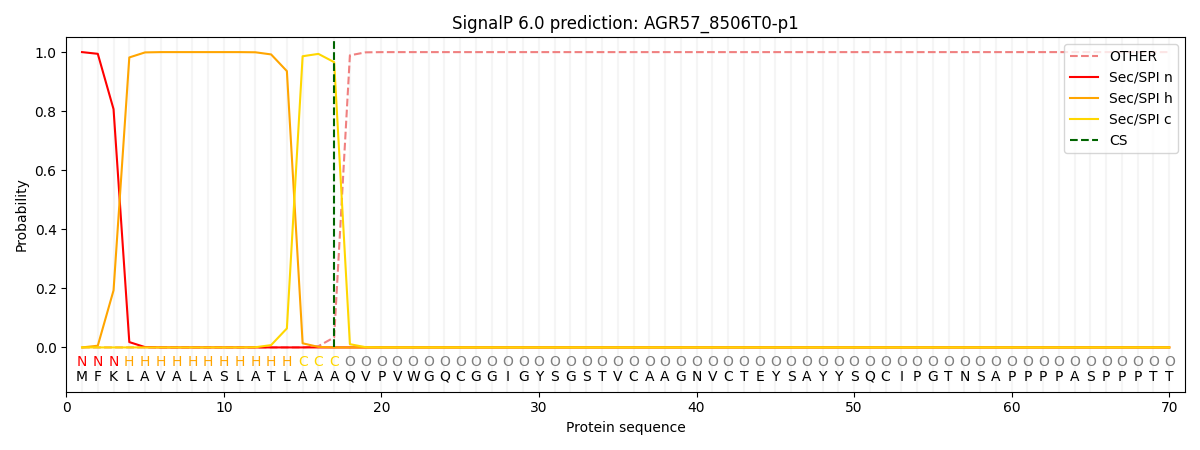
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000197 | 0.999775 | CS pos: 17-18. Pr: 0.9660 |
