You are browsing environment: FUNGIDB
CAZyme Information: AGR57_5877T0-p1
You are here: Home > Sequence: AGR57_5877T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phanerochaete chrysosporium | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Agaricomycetes; ; Phanerochaetaceae; Phanerochaete; Phanerochaete chrysosporium | |||||||||||
| CAZyme ID | AGR57_5877T0-p1 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | Lytic polysaccharide monooxygenase (formerly GH61) / Carbohydrate-Binding Module Family 1 protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA9 | 12 | 229 | 1.4e-61 | 0.9863636363636363 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 410622 | LPMO_AA9 | 9.64e-76 | 24 | 244 | 1 | 215 | lytic polysaccharide monooxygenase (LPMO) auxiliary activity family 9 (AA9). AA9 proteins are copper-dependent lytic polysaccharide monooxygenases (LPMOs) involved in the cleavage of cellulose chains with oxidation of carbons C1 and/or C4 and C6. Activities include lytic cellulose monooxygenase (C1-hydroxylating) (EC 1.14.99.54) and lytic cellulose monooxygenase (C4-dehydrogenating) (EC 1.14.99.56). The family used to be called GH61 because weak endoglucanase activity had been demonstrated in some family members. |
| 397484 | Glyco_hydro_61 | 7.30e-67 | 25 | 233 | 1 | 210 | Glycosyl hydrolase family 61. Although weak endoglucanase activity has been demonstrated in several members of this family, they lack the clustered conserved catalytic acidic amino acids present in most glycoside hydrolases. Many members of this family lack measurable cellulase activity on their own, but enhance the activity of other cellulolytic enzymes. They are therefore unlikely to be true glycoside hydrolases. The subsrate-binding surface of this family is a flat Ig-like fold. |
| 395595 | CBM_1 | 2.66e-13 | 297 | 325 | 1 | 29 | Fungal cellulose binding domain. |
| 197593 | fCBD | 1.31e-12 | 296 | 329 | 1 | 34 | Fungal-type cellulose-binding domain. Small four-cysteine cellulose-binding domain of fungi |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 5.35e-243 | 1 | 329 | 1 | 329 | |
| 1.28e-218 | 1 | 300 | 1 | 300 | |
| 1.27e-89 | 9 | 247 | 5 | 252 | |
| 1.75e-72 | 1 | 244 | 1 | 245 | |
| 4.01e-64 | 14 | 244 | 10 | 250 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.74e-41 | 24 | 241 | 1 | 216 | Chain A, LPMO9F [Malbranchea cinnamomea] |
|
| 9.52e-37 | 20 | 248 | 16 | 249 | Chain A, Cel61b [Trichoderma reesei],2VTC_B Chain B, Cel61b [Trichoderma reesei] |
|
| 7.39e-33 | 24 | 248 | 1 | 228 | Chain A, Lytic polysaccharide monooxygenase [Talaromyces verruculosus] |
|
| 2.50e-32 | 38 | 241 | 19 | 219 | Structural basis for substrate targeting and catalysis by fungal polysaccharide monooxygenases (PMO-3) [Neurospora crassa OR74A] |
|
| 6.19e-32 | 62 | 241 | 30 | 202 | Zinc-bound glycoside hydrolase 61 E from Thielavia terrestris [Thermothielavioides terrestris],3EII_B Zinc-bound glycoside hydrolase 61 E from Thielavia terrestris [Thermothielavioides terrestris],3EII_C Zinc-bound glycoside hydrolase 61 E from Thielavia terrestris [Thermothielavioides terrestris],3EII_D Zinc-bound glycoside hydrolase 61 E from Thielavia terrestris [Thermothielavioides terrestris],3EJA_A Magnesium-bound glycoside hydrolase 61 isoform E from Thielavia terrestris [Thermothielavioides terrestris],3EJA_B Magnesium-bound glycoside hydrolase 61 isoform E from Thielavia terrestris [Thermothielavioides terrestris],3EJA_C Magnesium-bound glycoside hydrolase 61 isoform E from Thielavia terrestris [Thermothielavioides terrestris],3EJA_D Magnesium-bound glycoside hydrolase 61 isoform E from Thielavia terrestris [Thermothielavioides terrestris] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.45e-46 | 21 | 329 | 17 | 342 | Polysaccharide monooxygenase Cel61a OS=Myceliophthora thermophila (strain ATCC 42464 / BCRC 31852 / DSM 1799) OX=573729 GN=Cel61a PE=1 SV=1 |
|
| 1.37e-41 | 11 | 329 | 7 | 347 | Probable endo-beta-1,4-glucanase D OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=eglD PE=3 SV=1 |
|
| 1.37e-41 | 11 | 329 | 7 | 347 | Probable endo-beta-1,4-glucanase D OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=eglD PE=3 SV=1 |
|
| 7.56e-40 | 11 | 329 | 7 | 346 | Probable endo-beta-1,4-glucanase D OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=eglD PE=3 SV=1 |
|
| 3.88e-38 | 11 | 329 | 7 | 358 | Probable endo-beta-1,4-glucanase D OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=eglD PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
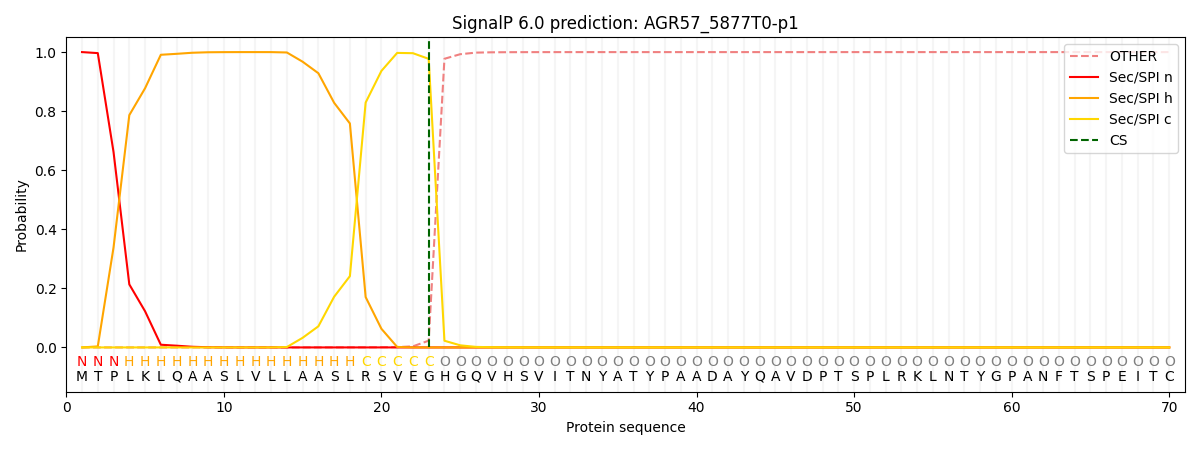
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000330 | 0.999673 | CS pos: 23-24. Pr: 0.9771 |
