You are browsing environment: FUNGIDB
CAZyme Information: AGR57_2006T0-p1
You are here: Home > Sequence: AGR57_2006T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phanerochaete chrysosporium | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Agaricomycetes; ; Phanerochaetaceae; Phanerochaete; Phanerochaete chrysosporium | |||||||||||
| CAZyme ID | AGR57_2006T0-p1 | |||||||||||
| CAZy Family | GH152 | |||||||||||
| CAZyme Description | Glycoside Hydrolase Family 47 protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.113:6 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH47 | 60 | 493 | 2.1e-137 | 0.9910313901345291 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396217 | Glyco_hydro_47 | 1.63e-145 | 60 | 492 | 3 | 451 | Glycosyl hydrolase family 47. Members of this family are alpha-mannosidases that catalyze the hydrolysis of the terminal 1,2-linked alpha-D-mannose residues in the oligo-mannose oligosaccharide Man(9)(GlcNAc)(2). |
| 240427 | PTZ00470 | 1.73e-72 | 56 | 490 | 77 | 514 | glycoside hydrolase family 47 protein; Provisional |
| 239047 | PA_GO-like | 0.003 | 705 | 760 | 18 | 75 | PA_GO-like: Protease-associated domain containing proteins like Arabidopsis thaliana growth-on protein GRO10. This group contains various PA domain-containing proteins similar to the functionally uncharacterized Arabidopsis GRO10. The PA domain may be a protein-protein interaction domain. At peptidase active sites, the PA domain may participate in substrate binding and/or promoting conformational changes, which influence the stability and accessibility of the site to substrate. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 8.51e-279 | 1 | 818 | 1 | 818 | |
| 5.34e-276 | 1 | 877 | 1 | 865 | |
| 1.63e-261 | 38 | 809 | 27 | 805 | |
| 1.15e-249 | 1 | 877 | 1 | 910 | |
| 8.29e-213 | 38 | 773 | 24 | 844 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.48e-52 | 57 | 492 | 11 | 449 | Crystal structure of the class I human endoplasmic reticulum 1,2-alpha-mannosidase and Man9GlcNAc2-PA complex [Homo sapiens] |
|
| 7.41e-52 | 57 | 492 | 16 | 454 | Crystal Structure Of Human Class I Alpha1,2-Mannosidase [Homo sapiens] |
|
| 7.74e-52 | 57 | 492 | 16 | 454 | Crystal Structure Of Human Class I Alpha1,2-Mannosidase In Complex With 1-Deoxymannojirimycin [Homo sapiens],1FO3_A Crystal Structure Of Human Class I Alpha1,2-Mannosidase In Complex With Kifunensine [Homo sapiens] |
|
| 3.30e-51 | 57 | 492 | 11 | 449 | Crystal structure of the class I human endoplasmic reticulum 1,2-alpha-mannosidase T688A mutant and Thio-disaccharide substrate analog complex [Homo sapiens],5KK7_B Crystal structure of the class I human endoplasmic reticulum 1,2-alpha-mannosidase T688A mutant and Thio-disaccharide substrate analog complex [Homo sapiens] |
|
| 3.77e-51 | 57 | 492 | 94 | 532 | Crystal Structure Of Human Class I alpha-1,2-Mannosidase In Complex With Thio-Disaccharide Substrate Analogue [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.92e-153 | 46 | 650 | 30 | 616 | ER degradation-enhancing alpha-mannosidase-like protein 1 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=mnl1 PE=3 SV=2 |
|
| 1.87e-144 | 56 | 500 | 130 | 587 | ER degradation-enhancing alpha-mannosidase-like protein 1 OS=Mus musculus OX=10090 GN=Edem1 PE=1 SV=1 |
|
| 6.63e-143 | 56 | 500 | 135 | 592 | ER degradation-enhancing alpha-mannosidase-like protein 1 OS=Homo sapiens OX=9606 GN=EDEM1 PE=1 SV=1 |
|
| 1.12e-122 | 40 | 490 | 26 | 474 | Alpha-mannosidase I MNS5 OS=Arabidopsis thaliana OX=3702 GN=MNS5 PE=1 SV=1 |
|
| 2.68e-121 | 56 | 481 | 40 | 452 | ER degradation-enhancing alpha-mannosidase-like protein 2 OS=Homo sapiens OX=9606 GN=EDEM2 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
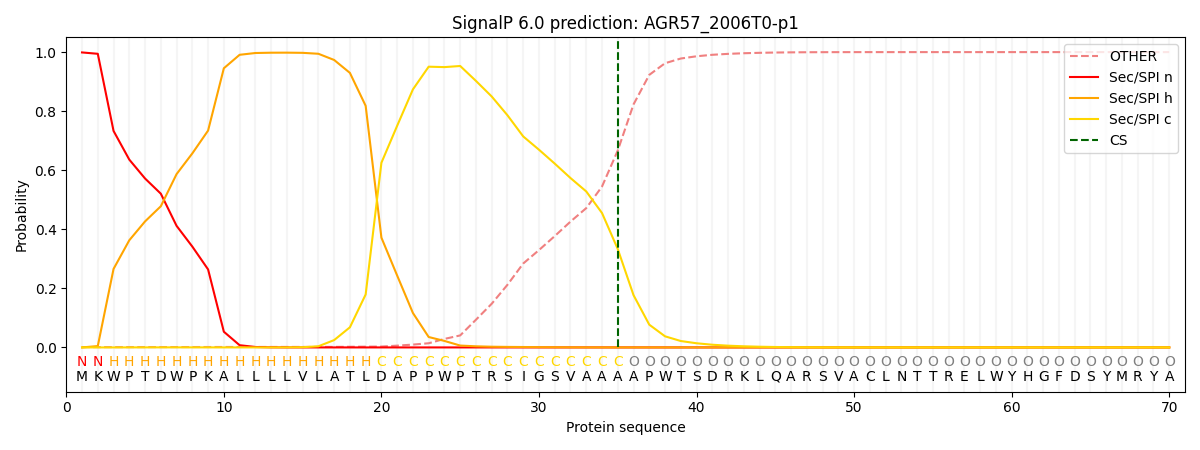
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.001873 | 0.998088 | CS pos: 35-36. Pr: 0.3332 |
