You are browsing environment: FUNGIDB
CAZyme Information: AGR57_13897T0-p1
You are here: Home > Sequence: AGR57_13897T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phanerochaete chrysosporium | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Agaricomycetes; ; Phanerochaetaceae; Phanerochaete; Phanerochaete chrysosporium | |||||||||||
| CAZyme ID | AGR57_13897T0-p1 | |||||||||||
| CAZy Family | CE1 | |||||||||||
| CAZyme Description | Carbohydrate-Binding Module Family 1 / Carbohydrate Esterase Family 15 protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.1.1.-:10 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE15 | 91 | 418 | 5.4e-87 | 0.9888475836431226 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395595 | CBM_1 | 1.99e-14 | 25 | 52 | 2 | 29 | Fungal cellulose binding domain. |
| 197593 | fCBD | 6.01e-13 | 24 | 56 | 2 | 34 | Fungal-type cellulose-binding domain. Small four-cysteine cellulose-binding domain of fungi |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.04e-217 | 9 | 452 | 8 | 474 | |
| 1.18e-179 | 76 | 452 | 4 | 376 | |
| 8.62e-169 | 77 | 452 | 31 | 405 | |
| 7.38e-155 | 20 | 452 | 19 | 466 | |
| 1.02e-152 | 13 | 452 | 10 | 457 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.34e-218 | 18 | 452 | 1 | 458 | Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor covalent complex with the aldouronic acid UXXR [Cerrena unicolor],6RV8_B Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor covalent complex with the aldouronic acid UXXR [Cerrena unicolor] |
|
| 3.40e-202 | 78 | 452 | 5 | 380 | Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor [Cerrena unicolor],6RU2_B Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor [Cerrena unicolor] |
|
| 9.04e-202 | 78 | 452 | 11 | 386 | Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant [Cerrena unicolor],6RTV_B Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant [Cerrena unicolor],6RU1_A Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant in complex with the aldouronic acid Um4X [Cerrena unicolor],6RU1_B Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant in complex with the aldouronic acid Um4X [Cerrena unicolor],6RV7_A Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant in complex with the aldouronic acid UXXR [Cerrena unicolor],6RV7_B Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant in complex with the aldouronic acid UXXR [Cerrena unicolor],6RV9_A Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant in complex with the aldouronic acid XUXXR [Cerrena unicolor],6RV9_B Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant in complex with the aldouronic acid XUXXR [Cerrena unicolor] |
|
| 1.88e-128 | 78 | 452 | 9 | 375 | Chain A, Cip2 [Trichoderma reesei],3PIC_B Chain B, Cip2 [Trichoderma reesei],3PIC_C Chain C, Cip2 [Trichoderma reesei] |
|
| 6.30e-106 | 77 | 438 | 45 | 400 | Crystal structure of recombinant glucuronoyl esterase from Sporotrichum thermophile determined at 1.55 A resolution [Thermothelomyces thermophilus ATCC 42464] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 0.0 | 1 | 452 | 1 | 472 | 4-O-methyl-glucuronoyl methylesterase 1 OS=Phanerochaete chrysosporium (strain RP-78 / ATCC MYA-4764 / FGSC 9002) OX=273507 GN=e_gw1.18.61.1 PE=1 SV=1 |
|
| 1.85e-218 | 9 | 452 | 8 | 474 | 4-O-methyl-glucuronoyl methylesterase OS=Cerrena unicolor OX=90312 PE=1 SV=1 |
|
| 8.33e-182 | 77 | 451 | 31 | 407 | 4-O-methyl-glucuronoyl methylesterase 1 OS=Wolfiporia cocos (strain MD-104) OX=742152 GN=WOLCODRAFT_23632 PE=1 SV=1 |
|
| 1.75e-177 | 77 | 452 | 22 | 393 | 4-O-methyl-glucuronoyl methylesterase OS=Schizophyllum commune (strain H4-8 / FGSC 9210) OX=578458 GN=SCHCODRAFT_238770 PE=1 SV=1 |
|
| 1.91e-175 | 77 | 449 | 31 | 402 | 4-O-methyl-glucuronoyl methylesterase 2 OS=Phanerochaete chrysosporium (strain RP-78 / ATCC MYA-4764 / FGSC 9002) OX=273507 GN=e_gw1.11.1537.1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
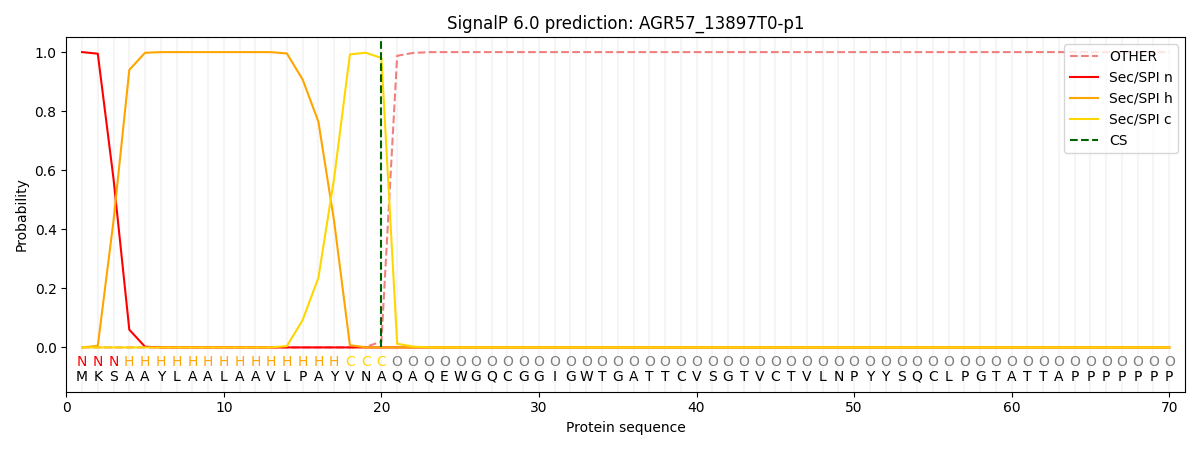
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000220 | 0.999763 | CS pos: 20-21. Pr: 0.9803 |
