You are browsing environment: FUNGIDB
CAZyme Information: AGR57_13663T0-p1
You are here: Home > Sequence: AGR57_13663T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phanerochaete chrysosporium | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Agaricomycetes; ; Phanerochaetaceae; Phanerochaete; Phanerochaete chrysosporium | |||||||||||
| CAZyme ID | AGR57_13663T0-p1 | |||||||||||
| CAZy Family | CE1 | |||||||||||
| CAZyme Description | Carbohydrate Esterase Family 8 protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 47 | 373 | 7.7e-58 | 0.9826388888888888 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 215367 | PLN02682 | 6.73e-34 | 41 | 380 | 67 | 367 | pectinesterase family protein |
| 395871 | Pectinesterase | 1.23e-32 | 47 | 375 | 3 | 296 | Pectinesterase. |
| 178087 | PLN02468 | 7.02e-30 | 47 | 380 | 261 | 554 | putative pectinesterase/pectinesterase inhibitor |
| 215383 | PLN02713 | 2.23e-28 | 166 | 375 | 342 | 549 | Probable pectinesterase/pectinesterase inhibitor |
| 178051 | PLN02432 | 6.05e-28 | 51 | 380 | 18 | 292 | putative pectinesterase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 9.75e-162 | 20 | 384 | 21 | 378 | |
| 9.75e-162 | 20 | 384 | 21 | 378 | |
| 2.78e-161 | 13 | 384 | 14 | 378 | |
| 1.95e-141 | 20 | 371 | 20 | 365 | |
| 1.90e-94 | 67 | 237 | 21 | 197 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.03e-19 | 42 | 380 | 5 | 308 | Pectin methylesterase from Carrot [Daucus carota] |
|
| 1.15e-14 | 200 | 375 | 125 | 300 | Chain A, Pectinesterase 1 [Solanum lycopersicum] |
|
| 1.45e-07 | 48 | 346 | 34 | 328 | The structure of rice weevil pectin methyl esterase [Sitophilus oryzae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.26e-24 | 51 | 375 | 264 | 556 | Probable pectinesterase/pectinesterase inhibitor 41 OS=Arabidopsis thaliana OX=3702 GN=PME41 PE=2 SV=2 |
|
| 2.18e-23 | 47 | 380 | 223 | 525 | Probable pectinesterase/pectinesterase inhibitor 59 OS=Arabidopsis thaliana OX=3702 GN=PME59 PE=2 SV=1 |
|
| 4.81e-22 | 116 | 380 | 287 | 553 | Probable pectinesterase/pectinesterase inhibitor 46 OS=Arabidopsis thaliana OX=3702 GN=PME46 PE=2 SV=1 |
|
| 1.17e-21 | 41 | 384 | 32 | 324 | Probable pectinesterase A OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=pmeA PE=3 SV=1 |
|
| 1.32e-21 | 166 | 380 | 393 | 604 | Probable pectinesterase/pectinesterase inhibitor 25 OS=Arabidopsis thaliana OX=3702 GN=PME25 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
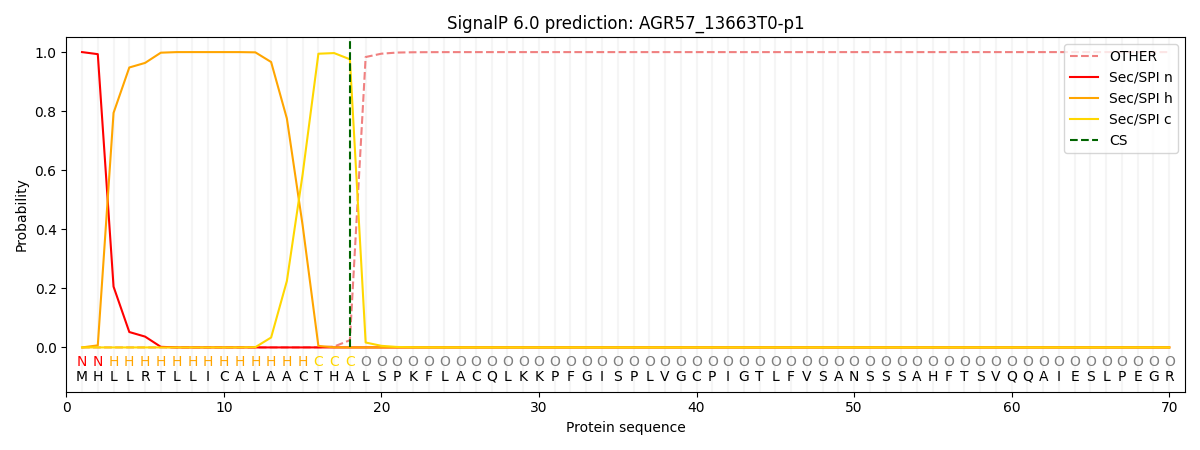
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000236 | 0.999761 | CS pos: 18-19. Pr: 0.9757 |
