You are browsing environment: FUNGIDB
CAZyme Information: AGR57_12035T0-p1
You are here: Home > Sequence: AGR57_12035T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phanerochaete chrysosporium | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Agaricomycetes; ; Phanerochaetaceae; Phanerochaete; Phanerochaete chrysosporium | |||||||||||
| CAZyme ID | AGR57_12035T0-p1 | |||||||||||
| CAZy Family | AA3 | |||||||||||
| CAZyme Description | Glycosyltransferase Family 22 protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT22 | 65 | 547 | 2.1e-90 | 0.9897172236503856 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 281842 | Glyco_transf_22 | 1.23e-77 | 63 | 548 | 2 | 414 | Alg9-like mannosyltransferase family. Members of this family are mannosyltransferase enzymes. At least some members are localized in endoplasmic reticulum and involved in GPI anchor biosynthesis. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 9.06e-231 | 8 | 686 | 4 | 679 | |
| 6.06e-218 | 9 | 686 | 5 | 698 | |
| 1.45e-196 | 6 | 676 | 1 | 682 | |
| 1.16e-195 | 6 | 682 | 1 | 688 | |
| 2.88e-195 | 10 | 679 | 10 | 678 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.27e-116 | 56 | 679 | 54 | 600 | Alpha-1,2-mannosyltransferase ALG9 OS=Mus musculus OX=10090 GN=Alg9 PE=2 SV=1 |
|
| 9.39e-113 | 56 | 679 | 54 | 600 | Alpha-1,2-mannosyltransferase ALG9 OS=Homo sapiens OX=9606 GN=ALG9 PE=1 SV=2 |
|
| 1.23e-96 | 64 | 660 | 18 | 545 | Alpha-1,2-mannosyltransferase alg9 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=alg9 PE=3 SV=1 |
|
| 1.37e-80 | 56 | 660 | 49 | 583 | Alpha-1,2-mannosyltransferase algn-9 OS=Caenorhabditis elegans OX=6239 GN=algn-9 PE=1 SV=2 |
|
| 5.52e-71 | 68 | 675 | 11 | 546 | Alpha-1,2-mannosyltransferase ALG9 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=ALG9 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
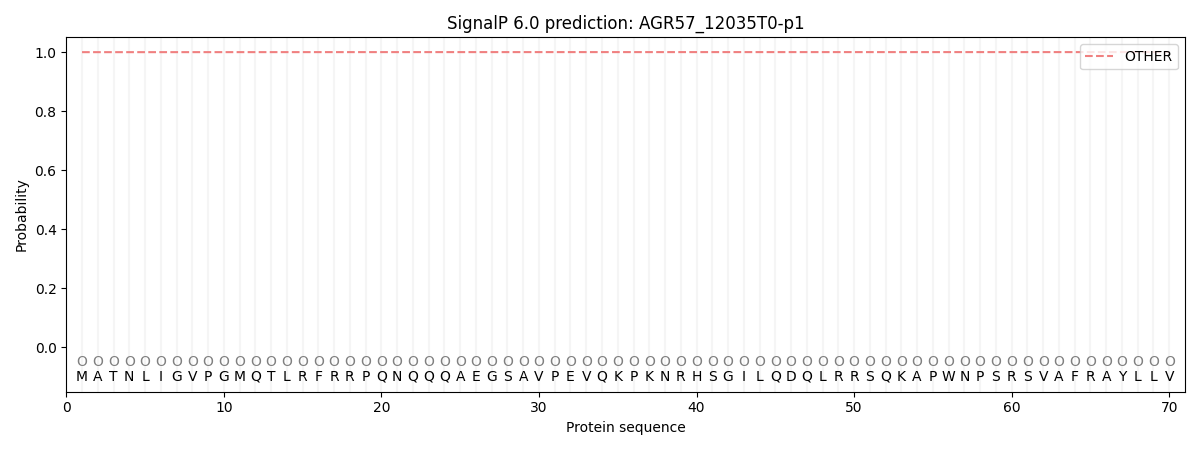
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999846 | 0.000169 |
TMHMM Annotations download full data without filtering help
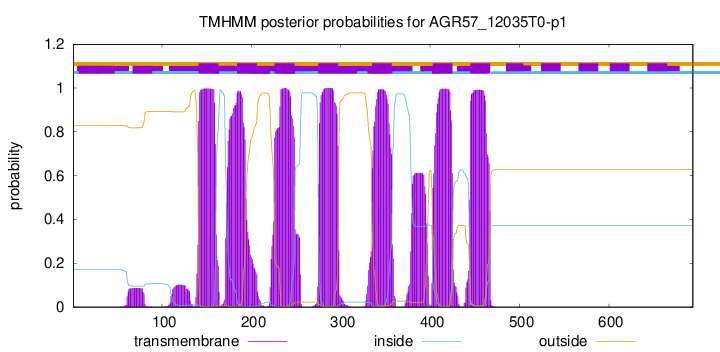
| Start | End |
|---|---|
| 141 | 163 |
| 184 | 206 |
| 226 | 248 |
| 275 | 297 |
| 335 | 357 |
| 403 | 425 |
| 445 | 467 |
