You are browsing environment: FUNGIDB
CAZyme Information: AGR57_10960T0-p1
You are here: Home > Sequence: AGR57_10960T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phanerochaete chrysosporium | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Agaricomycetes; ; Phanerochaetaceae; Phanerochaete; Phanerochaete chrysosporium | |||||||||||
| CAZyme ID | AGR57_10960T0-p1 | |||||||||||
| CAZy Family | AA3 | |||||||||||
| CAZyme Description | Glycoside Hydrolase Family 74 / Carbohydrate-Binding Module Family 1 protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.151:27 | 3.2.1.150:4 | 3.2.1.151:5 | 3.2.1.151:27 | 3.2.1.150:4 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH74 | 79 | 189 | 2e-24 | 0.4592274678111588 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 214740 | VPS10 | 2.20e-06 | 117 | 201 | 55 | 136 | VPS10 domain. |
| 395595 | CBM_1 | 8.98e-06 | 833 | 849 | 1 | 17 | Fungal cellulose binding domain. |
| 406353 | Sortilin-Vps10 | 9.65e-06 | 70 | 184 | 12 | 112 | Sortilin, neurotensin receptor 3,. Sortilin, also known in mammals as neurotensin receptor-3, is the archetypical member of a Vps10-domain (Vps10-D) that binds neurotrophic factors and neuropeptides. This domain constitutes the entire luminal part of Sortilin and is activated in the trans-Golgi network by enzymatic propeptide cleavage. The structure of the domain has been determined as a ten-bladed propeller, with up to 9 BNR or beta-hairpin turns in it. The mature receptor binds various ligands, including its own propeptide (Sort-pro), neurotensin, the pro-forms of nerve growth factor-beta (NGF)6 and brain-derived neurotrophic factor (BDNF)7, lipoprotein lipase (LpL), apo lipoprotein AV14 and the receptor-associated protein (RAP)1. |
| 271234 | Sialidase_non-viral | 4.20e-04 | 100 | 294 | 157 | 322 | Non-viral sialidases. Sialidases or neuraminidases function to bind and hydrolyze terminal sialic acid residues from various glycoconjugates, they play vital roles in pathogenesis, bacterial nutrition and cellular interactions. They have a six-bladed, beta-propeller fold with the non-viral sialidases containing 2-5 Asp-box motifs (most commonly Ser/Thr-X-Asp-[X]-Gly-X-Thr- Trp/Phe). This CD includes eubacterial and eukaryotic sialidases. |
| 197593 | fCBD | 0.002 | 833 | 853 | 2 | 22 | Fungal-type cellulose-binding domain. Small four-cysteine cellulose-binding domain of fungi |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 21 | 887 | 1 | 854 | |
| 0.0 | 5 | 848 | 5 | 822 | |
| 0.0 | 5 | 848 | 5 | 822 | |
| 0.0 | 12 | 848 | 13 | 820 | |
| 0.0 | 24 | 763 | 24 | 752 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.18e-232 | 21 | 757 | 2 | 737 | The structure of Acidothermus cellulolyticus family 74 glycoside hydrolase [Acidothermus cellulolyticus 11B] |
|
| 5.27e-225 | 24 | 754 | 4 | 745 | Crystal structure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, D60A mutant in complex with XXLG and XGXXLG xyloglucan [Paenibacillus odorifer] |
|
| 1.09e-224 | 21 | 754 | 1 | 746 | Crystal structure of Paenibacillus graminis GH74 (PgGH74) [Paenibacillus graminis] |
|
| 1.73e-220 | 24 | 754 | 4 | 745 | Crystal structure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, apoenzyme [Paenibacillus odorifer],6MGJ_B Crystal structure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, apoenzyme [Paenibacillus odorifer],6MGJ_C Crystal structure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, apoenzyme [Paenibacillus odorifer],6MGJ_D Crystal structure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, apoenzyme [Paenibacillus odorifer],6MGJ_E Crystal structure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, apoenzyme [Paenibacillus odorifer],6MGJ_F Crystal structure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, apoenzyme [Paenibacillus odorifer],6MGJ_G Crystal structure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, apoenzyme [Paenibacillus odorifer],6MGJ_H Crystal structure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, apoenzyme [Paenibacillus odorifer],6MGK_A Crystal structure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, in complex with XLX xyloglucan [Paenibacillus odorifer],6MGK_B Crystal structure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, in complex with XLX xyloglucan [Paenibacillus odorifer],6MGK_C Crystal structure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, in complex with XLX xyloglucan [Paenibacillus odorifer],6MGK_D Crystal structure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, in complex with XLX xyloglucan [Paenibacillus odorifer] |
|
| 2.36e-212 | 24 | 754 | 1 | 725 | Crystal structure of Streptomyces rapamycinicus GH74 in complex with xyloglucan fragments XLLG and XXXG [Streptomyces rapamycinicus NRRL 5491],6P2O_B Crystal structure of Streptomyces rapamycinicus GH74 in complex with xyloglucan fragments XLLG and XXXG [Streptomyces rapamycinicus NRRL 5491] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.48e-273 | 9 | 752 | 8 | 742 | Xyloglucanase OS=Hypocrea jecorina (strain QM6a) OX=431241 GN=cel74a PE=1 SV=1 |
|
| 3.45e-230 | 20 | 768 | 32 | 784 | Xyloglucanase OS=Paenibacillus sp. OX=58172 GN=xeg74 PE=1 SV=1 |
|
| 3.93e-203 | 7 | 763 | 14 | 766 | Xyloglucanase Xgh74A OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=xghA PE=3 SV=1 |
|
| 1.56e-202 | 7 | 763 | 14 | 766 | Xyloglucanase Xgh74A OS=Acetivibrio thermocellus OX=1515 GN=xghA PE=1 SV=1 |
|
| 1.63e-106 | 4 | 754 | 11 | 784 | Oligoxyloglucan reducing end-specific cellobiohydrolase OS=Geotrichum sp. (strain M128) OX=203496 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
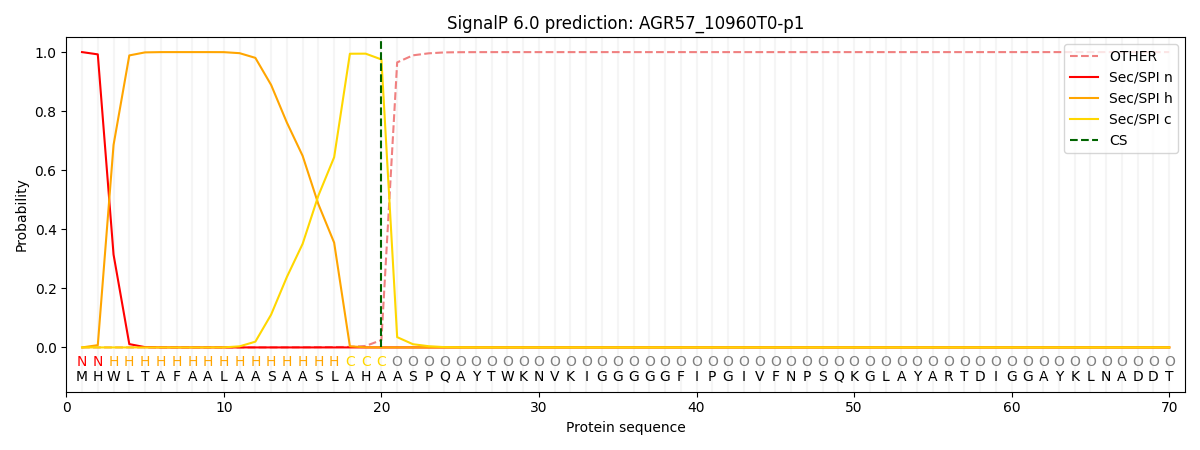
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000232 | 0.999750 | CS pos: 20-21. Pr: 0.9752 |
