You are browsing environment: FUNGIDB
CAZyme Information: AGR57_10863T0-p1
Basic Information
help
| Species |
Phanerochaete chrysosporium
|
| Lineage |
Basidiomycota; Agaricomycetes; ; Phanerochaetaceae; Phanerochaete; Phanerochaete chrysosporium
|
| CAZyme ID |
AGR57_10863T0-p1
|
| CAZy Family |
PL42 |
| CAZyme Description |
Glycosyltransferase Family 31 protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 650 |
|
73373.03 |
8.0898 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_PchrysosporiumRP-78 |
13602 |
273507 |
0 |
13602
|
|
| Gene Location |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 215596
|
PLN03133 |
0.002 |
203 |
280 |
384 |
451 |
beta-1,3-galactosyltransferase; Provisional |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 1.64e-152 |
77 |
645 |
455 |
957 |
| 1.94e-118 |
123 |
642 |
164 |
788 |
| 4.90e-117 |
57 |
642 |
96 |
799 |
| 3.84e-115 |
57 |
639 |
96 |
792 |
| 1.08e-113 |
57 |
639 |
96 |
790 |
AGR57_10863T0-p1 has no PDB hit.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 8.91e-12 |
203 |
370 |
63 |
221 |
Beta-1,3-galactosyltransferase pvg3 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=pvg3 PE=1 SV=1 |
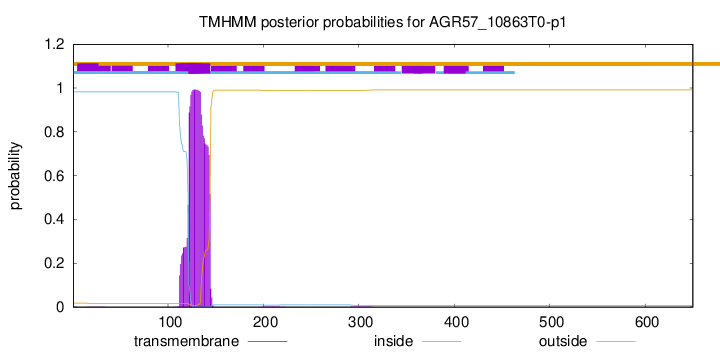
This protein is predicted as OTHER
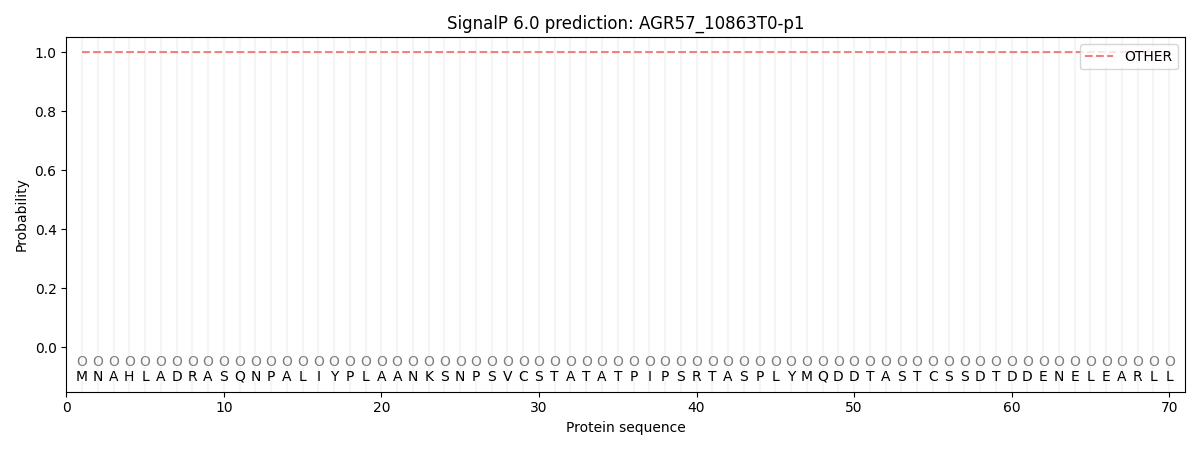
| Other |
SP_Sec_SPI |
CS Position |
| 1.000070 |
0.000001 |
|