You are browsing environment: FUNGIDB
CAZyme Information: AGR57_10553T0-p1
Basic Information
help
| Species |
Phanerochaete chrysosporium
|
| Lineage |
Basidiomycota; Agaricomycetes; ; Phanerochaetaceae; Phanerochaete; Phanerochaete chrysosporium
|
| CAZyme ID |
AGR57_10553T0-p1
|
| CAZy Family |
AA2 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_PchrysosporiumRP-78 |
13602 |
273507 |
0 |
13602
|
|
| Gene Location |
No EC number prediction in AGR57_10553T0-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| PL35 |
519 |
707 |
5.4e-52 |
0.9832402234636871 |
AGR57_10553T0-p1 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 0.0 |
10 |
785 |
19 |
790 |
| 0.0 |
10 |
785 |
19 |
790 |
| 0.0 |
10 |
785 |
20 |
799 |
| 1.27e-274 |
134 |
785 |
46 |
696 |
| 7.94e-262 |
104 |
785 |
129 |
812 |
AGR57_10553T0-p1 has no PDB hit.
AGR57_10553T0-p1 has no Swissprot hit.
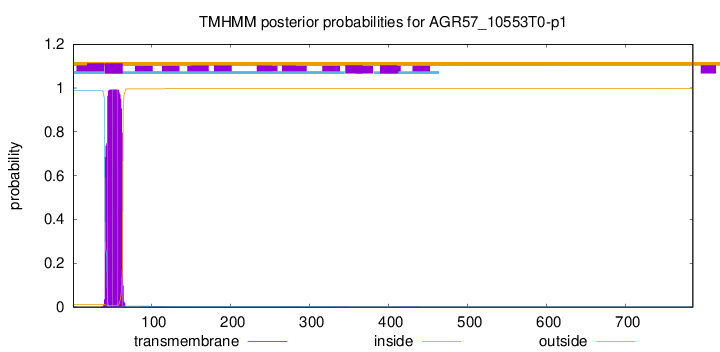
This protein is predicted as OTHER
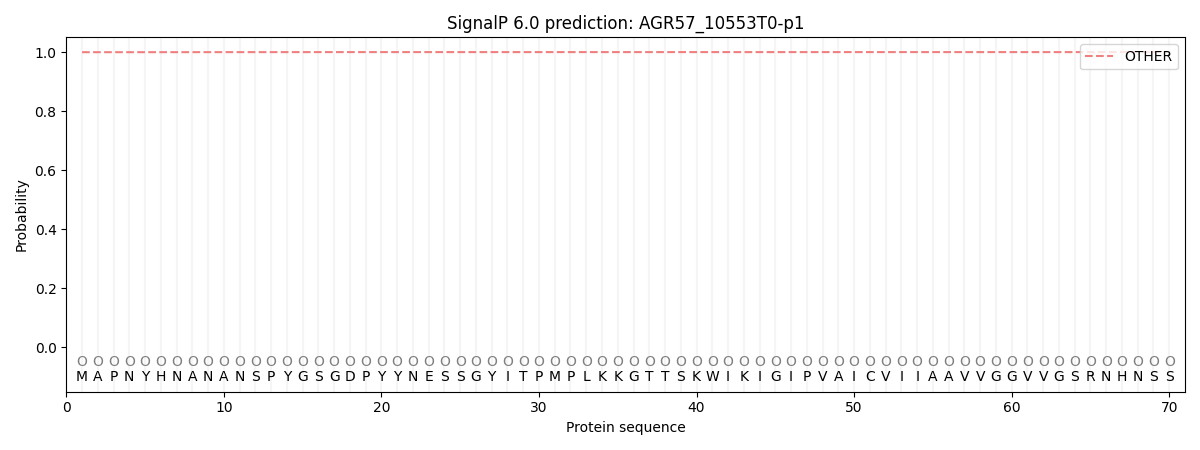
| Other |
SP_Sec_SPI |
CS Position |
| 0.999828 |
0.000186 |
|