You are browsing environment: FUNGIDB
CAZyme Information: AFUB_096220-T-p1
Basic Information
help
| Species |
Aspergillus fumigatus
|
| Lineage |
Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus fumigatus
|
| CAZyme ID |
AFUB_096220-T-p1
|
| CAZy Family |
PL4 |
| CAZyme Description |
Ortholog(s) have UDP-galactosyltransferase activity, role in beta-D-galactofuranose biosynthetic process and Golgi apparatus localization
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 537 |
|
61284.55 |
5.9050 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_AfumigatusA1163 |
10106 |
451804 |
199 |
9907
|
|
| Gene Location |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 215605
|
PLN03153 |
4.63e-10 |
244 |
407 |
202 |
375 |
hypothetical protein; Provisional |
| 398362
|
DUF604 |
0.001 |
303 |
397 |
10 |
106 |
Protein of unknown function, DUF604. This family includes a conserved region found in several uncharacterized plant proteins. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 0.0 |
1 |
537 |
1 |
537 |
| 0.0 |
4 |
537 |
3 |
537 |
| 0.0 |
4 |
537 |
3 |
537 |
| 0.0 |
4 |
537 |
3 |
537 |
| 0.0 |
14 |
537 |
1 |
525 |
AFUB_096220-T-p1 has no PDB hit.
AFUB_096220-T-p1 has no Swissprot hit.
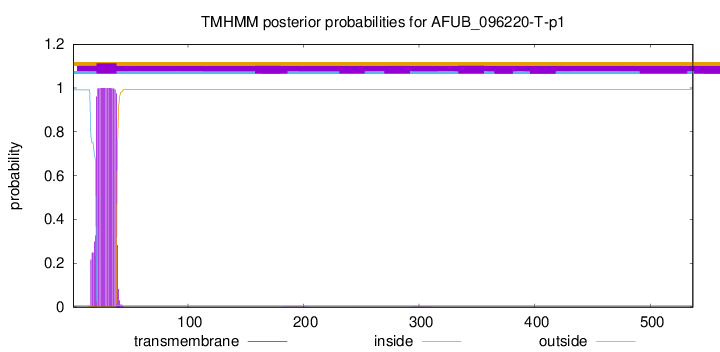
This protein is predicted as OTHER

| Other |
SP_Sec_SPI |
CS Position |
| 0.999951 |
0.000065 |
|