You are browsing environment: FUNGIDB
CAZyme Information: AFUB_095140-T-p1
Basic Information
help
| Species |
Aspergillus fumigatus
|
| Lineage |
Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus fumigatus
|
| CAZyme ID |
AFUB_095140-T-p1
|
| CAZy Family |
PL20 |
| CAZyme Description |
Has domain(s) with predicted cation binding, hydrolase activity, hydrolyzing O-glycosyl compounds activity and role in carbohydrate metabolic process
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 430 |
|
47442.89 |
6.2233 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_AfumigatusA1163 |
10106 |
451804 |
199 |
9907
|
|
| Gene Location |
No EC number prediction in AFUB_095140-T-p1.
AFUB_095140-T-p1 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 2.83e-235 |
1 |
430 |
1 |
483 |
| 2.93e-197 |
6 |
430 |
5 |
463 |
| 5.89e-197 |
8 |
430 |
7 |
463 |
| 7.90e-188 |
26 |
428 |
18 |
453 |
| 3.06e-143 |
25 |
429 |
19 |
455 |
AFUB_095140-T-p1 has no PDB hit.
AFUB_095140-T-p1 has no Swissprot hit.
This protein is predicted as SP
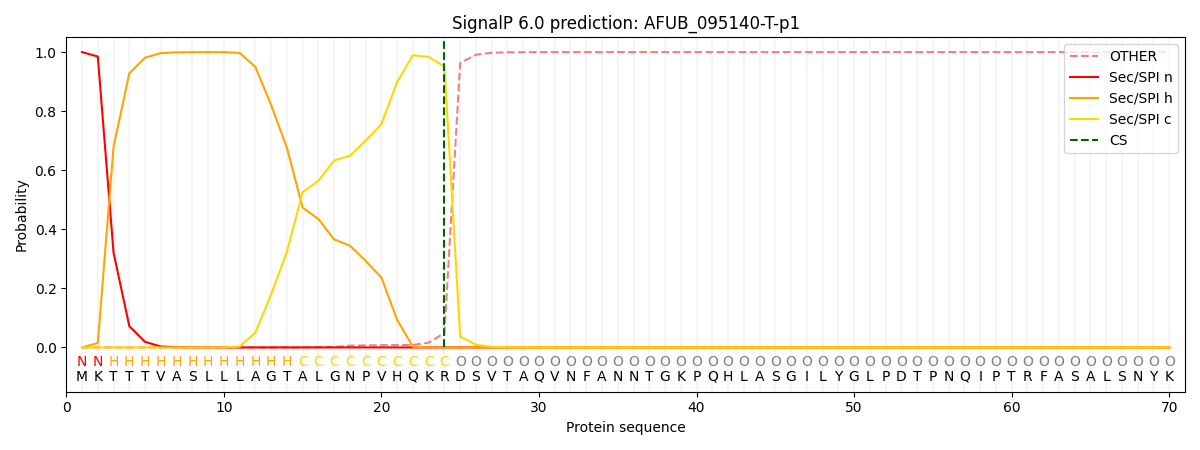
| Other |
SP_Sec_SPI |
CS Position |
| 0.000436 |
0.999522 |
CS pos: 24-25. Pr: 0.9512 |
There is no transmembrane helices in AFUB_095140-T-p1.

