You are browsing environment: FUNGIDB
CAZyme Information: AEO61799.1
You are here: Home > Sequence: AEO61799.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Thermothelomyces thermophilus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Chaetomiaceae; Thermothelomyces; Thermothelomyces thermophilus | |||||||||||
| CAZyme ID | AEO61799.1 | |||||||||||
| CAZy Family | GT76 | |||||||||||
| CAZyme Description | carbohydrate esterase family 1 protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 226040 | LpqC | 2.30e-05 | 92 | 243 | 119 | 278 | Poly(3-hydroxybutyrate) depolymerase [Secondary metabolites biosynthesis, transport and catabolism]. |
| 402228 | Esterase_phd | 1.01e-04 | 90 | 197 | 70 | 194 | Esterase PHB depolymerase. This family of proteins include acetyl xylan esterases (AXE), feruloyl esterases (FAE), and poly(3-hydroxybutyrate) (PHB) depolymerases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 8.41e-182 | 1 | 254 | 1 | 254 | |
| 2.93e-84 | 14 | 254 | 12 | 286 | |
| 4.76e-83 | 14 | 254 | 12 | 286 | |
| 4.76e-83 | 14 | 254 | 12 | 286 | |
| 5.22e-69 | 14 | 254 | 9 | 296 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.56e-48 | 30 | 254 | 9 | 274 | Acetyl xylan esterase from Aspergillus awamori [Aspergillus awamori],5X6S_B Acetyl xylan esterase from Aspergillus awamori [Aspergillus awamori] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.77e-51 | 41 | 254 | 51 | 306 | Probable acetylxylan esterase A OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=axeA PE=3 SV=1 |
|
| 1.07e-49 | 30 | 254 | 37 | 302 | Acetylxylan esterase A OS=Aspergillus ficuum OX=5058 GN=axeA PE=1 SV=1 |
|
| 5.41e-49 | 41 | 254 | 49 | 304 | Probable acetylxylan esterase A OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=axeA PE=3 SV=1 |
|
| 7.34e-49 | 30 | 254 | 40 | 310 | Probable acetylxylan esterase A OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=axeA PE=3 SV=1 |
|
| 8.40e-49 | 30 | 254 | 37 | 302 | Probable acetylxylan esterase A OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=axeA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
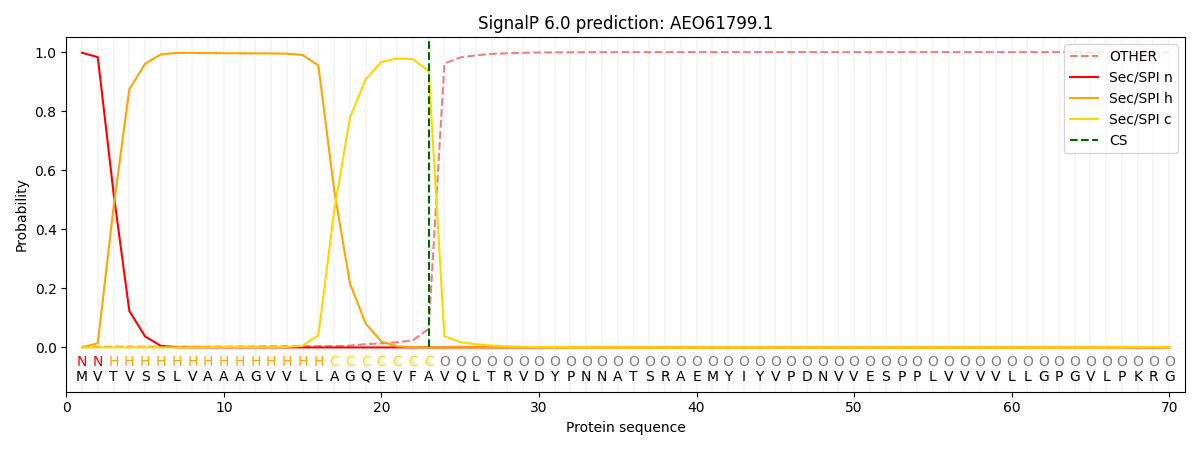
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.010854 | 0.989130 | CS pos: 23-24. Pr: 0.9370 |
