You are browsing environment: FUNGIDB
CAZyme Information: AEO60897.1
You are here: Home > Sequence: AEO60897.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Thermothelomyces thermophilus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Chaetomiaceae; Thermothelomyces; Thermothelomyces thermophilus | |||||||||||
| CAZyme ID | AEO60897.1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | glycoside hydrolase family 5 protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.78:33 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 58 | 330 | 2.3e-85 | 0.9930795847750865 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 226444 | COG3934 | 2.23e-31 | 34 | 360 | 4 | 305 | Endo-1,4-beta-mannosidase [Carbohydrate transport and metabolism]. |
| 395098 | Cellulase | 4.75e-17 | 37 | 332 | 4 | 270 | Cellulase (glycosyl hydrolase family 5). |
| 225344 | BglC | 1.97e-05 | 56 | 298 | 71 | 304 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 6.68e-285 | 1 | 370 | 1 | 370 | |
| 3.41e-225 | 1 | 367 | 1 | 365 | |
| 3.83e-206 | 1 | 365 | 2 | 367 | |
| 2.10e-204 | 1 | 365 | 1 | 369 | |
| 8.52e-204 | 1 | 365 | 1 | 369 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.13e-200 | 18 | 365 | 4 | 355 | Chain A, Gh5 Endo-beta-1,4-mannanase [Podospora anserina] |
|
| 7.86e-168 | 30 | 365 | 8 | 344 | Chain A, ENDO-1,4-B-D-MANNANASE [Trichoderma reesei],1QNP_A Chain A, Endo-1,4-b-d-mannanase [Trichoderma reesei],1QNQ_A Chain A, Endo-1,4-b-d-mannanase [Trichoderma reesei],1QNR_A Chain A, Endo-1,4-b-d-mannanase [Trichoderma reesei],1QNS_A Chain A, Endo-1,4-b-d-mannanase [Trichoderma reesei] |
|
| 2.36e-155 | 30 | 367 | 6 | 345 | The ligand-free structure of ManBK from Aspergillus niger BK01 [Aspergillus niger],3WH9_B The ligand-free structure of ManBK from Aspergillus niger BK01 [Aspergillus niger] |
|
| 1.19e-147 | 28 | 364 | 5 | 342 | Crtstal structure of glycoside hydrolase family 5 beta-mannanase from Talaromyces trachyspermus [Talaromyces trachyspermus] |
|
| 1.34e-72 | 30 | 364 | 10 | 381 | The Crystal Structure of Chrysonilia sitophila endo-beta-D-1,4- mannanase [Neurospora sitophila] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.51e-204 | 1 | 365 | 1 | 369 | Mannan endo-1,4-beta-mannosidase A OS=Podospora anserina (strain S / ATCC MYA-4624 / DSM 980 / FGSC 10383) OX=515849 GN=Pa_6_490 PE=1 SV=1 |
|
| 3.22e-167 | 14 | 366 | 19 | 372 | Mannan endo-1,4-beta-mannosidase A OS=Hypocrea jecorina (strain ATCC 56765 / BCRC 32924 / NRRL 11460 / Rut C-30) OX=1344414 GN=man1 PE=1 SV=1 |
|
| 4.18e-165 | 19 | 365 | 34 | 383 | Probable mannan endo-1,4-beta-mannosidase A OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=manA PE=3 SV=1 |
|
| 2.48e-164 | 20 | 365 | 26 | 373 | Mannan endo-1,4-beta-mannosidase A OS=Aspergillus aculeatus OX=5053 GN=manA PE=1 SV=1 |
|
| 2.78e-163 | 19 | 365 | 34 | 383 | Probable mannan endo-1,4-beta-mannosidase A OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=manA PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
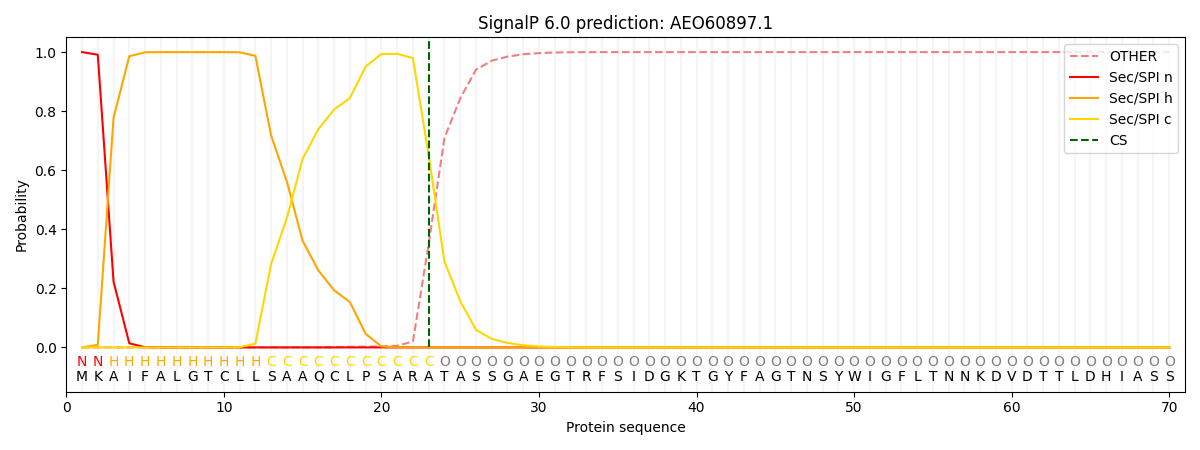
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000277 | 0.999697 | CS pos: 23-24. Pr: 0.6494 |
