You are browsing environment: FUNGIDB
CAZyme Information: AEO58960.1
You are here: Home > Sequence: AEO58960.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Thermothelomyces thermophilus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Chaetomiaceae; Thermothelomyces; Thermothelomyces thermophilus | |||||||||||
| CAZyme ID | AEO58960.1 | |||||||||||
| CAZy Family | GH43 | |||||||||||
| CAZyme Description | glycoside hydrolase family 33 protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.18:1 | 3.2.1.-:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH33 | 53 | 396 | 3.2e-55 | 0.9444444444444444 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 271229 | Sialidase | 5.22e-93 | 35 | 399 | 1 | 361 | sialidases/neuraminidases. Sialidases or neuraminidases function to bind and hydrolyze terminal sialic acid residues from various glycoconjugates as well as playing roles in pathogenesis, bacterial nutrition and cellular interactions. They have a six-bladed beta-propeller fold. This hierarchy includes eubacterial, eukaryotic, and viral sialidases. |
| 271234 | Sialidase_non-viral | 9.74e-60 | 54 | 399 | 10 | 339 | Non-viral sialidases. Sialidases or neuraminidases function to bind and hydrolyze terminal sialic acid residues from various glycoconjugates, they play vital roles in pathogenesis, bacterial nutrition and cellular interactions. They have a six-bladed, beta-propeller fold with the non-viral sialidases containing 2-5 Asp-box motifs (most commonly Ser/Thr-X-Asp-[X]-Gly-X-Thr- Trp/Phe). This CD includes eubacterial and eukaryotic sialidases. |
| 404074 | BNR_2 | 3.05e-07 | 69 | 275 | 1 | 188 | BNR repeat-like domain. This family of proteins contains BNR-like repeats suggesting these proteins may act as sialidases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 9.31e-314 | 1 | 409 | 1 | 409 | |
| 9.60e-226 | 21 | 409 | 21 | 406 | |
| 7.87e-220 | 33 | 409 | 2 | 376 | |
| 8.27e-208 | 2 | 409 | 14 | 424 | |
| 1.69e-190 | 1 | 409 | 1 | 403 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.59e-227 | 21 | 409 | 1 | 386 | The Aspergillus Fumigatus Sialidase Is A Kdnase: Structural And Mechanistic Insights [Aspergillus fumigatus Af293],2XZI_B The Aspergillus Fumigatus Sialidase Is A Kdnase: Structural And Mechanistic Insights [Aspergillus fumigatus Af293],2XZJ_A The Aspergillus Fumigatus Sialidase Is A Kdnase: Structural And Mechanistic Insights [Aspergillus fumigatus Af293],2XZJ_B The Aspergillus Fumigatus Sialidase Is A Kdnase: Structural And Mechanistic Insights [Aspergillus fumigatus Af293],2XZK_A The Aspergillus Fumigatus Sialidase Is A Kdnase: Structural And Mechanistic Insights [Aspergillus fumigatus Af293],2XZK_B The Aspergillus Fumigatus Sialidase Is A Kdnase: Structural And Mechanistic Insights [Aspergillus fumigatus Af293] |
|
| 3.44e-226 | 21 | 409 | 37 | 422 | Structural evaluation the Y358H mutant of the aspergillus fumigatus kdnase (sialidase) [Aspergillus fumigatus Af293],4M4N_B Structural evaluation the Y358H mutant of the aspergillus fumigatus kdnase (sialidase) [Aspergillus fumigatus Af293] |
|
| 6.93e-226 | 21 | 409 | 37 | 422 | Structural evaluation R171L mutant of the aspergillus fumigatus kdnase (sialidase) [Aspergillus fumigatus Af293],4M4V_B Structural evaluation R171L mutant of the aspergillus fumigatus kdnase (sialidase) [Aspergillus fumigatus Af293] |
|
| 9.83e-226 | 21 | 409 | 37 | 422 | Structural evaluation D84A mutant of the aspergillus fumigatus kdnase (sialidase) [Aspergillus fumigatus Af293],4M4U_B Structural evaluation D84A mutant of the aspergillus fumigatus kdnase (sialidase) [Aspergillus fumigatus Af293] |
|
| 1.24e-224 | 21 | 409 | 1 | 386 | Crystal structure of Aspergillus fumigatus sialidase [Aspergillus fumigatus],2XCY_B Crystal structure of Aspergillus fumigatus sialidase [Aspergillus fumigatus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.71e-226 | 21 | 409 | 21 | 406 | Exo-alpha-sialidase OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=AFUA_4G13800 PE=1 SV=2 |
|
| 1.93e-221 | 2 | 409 | 5 | 408 | Exo-alpha-sialidase ARB_03431 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_03431 PE=1 SV=1 |
|
| 2.19e-15 | 54 | 403 | 63 | 401 | Sialidase OS=Micromonospora viridifaciens OX=1881 GN=nedA PE=1 SV=1 |
|
| 1.84e-11 | 57 | 378 | 202 | 527 | Sialidase OS=Bacteroides fragilis (strain YCH46) OX=295405 GN=nanH PE=3 SV=2 |
|
| 5.82e-09 | 57 | 221 | 23 | 178 | Sialidase-3 OS=Bos taurus OX=9913 GN=NEU3 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
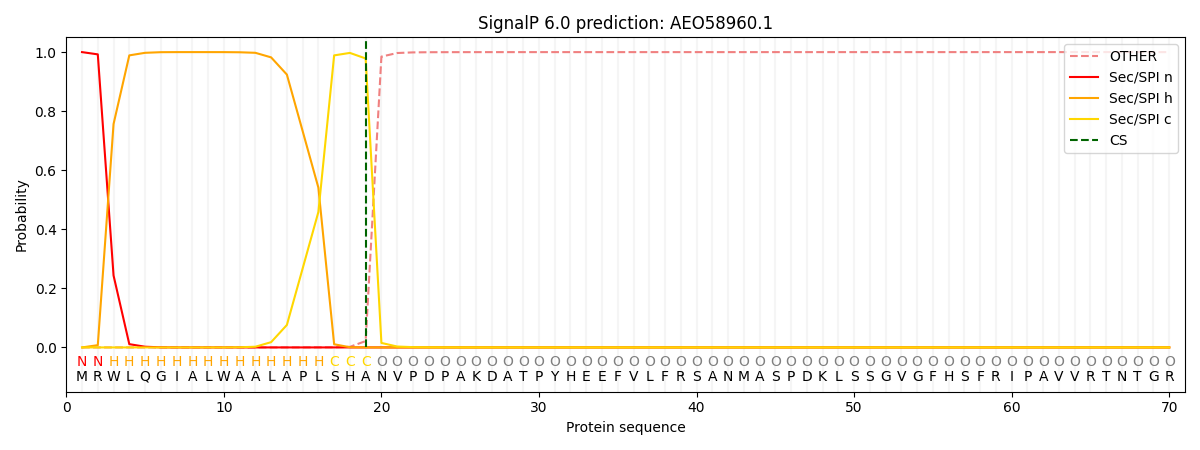
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000287 | 0.999681 | CS pos: 19-20. Pr: 0.9784 |
