You are browsing environment: FUNGIDB
CAZyme Information: ACLA_097280-t26_1-p1
Basic Information
help
| Species |
Aspergillus clavatus
|
| Lineage |
Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus clavatus
|
| CAZyme ID |
ACLA_097280-t26_1-p1
|
| CAZy Family |
PL20 |
| CAZyme Description |
conserved hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 462 |
DS027058|CGC8 |
51107.78 |
6.0998 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_AclavatusNRRL1 |
9428 |
344612 |
287 |
9141
|
|
| Gene Location |
No EC number prediction in ACLA_097280-t26_1-p1.
ACLA_097280-t26_1-p1 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 2.58e-265 |
3 |
462 |
4 |
483 |
| 2.20e-217 |
27 |
461 |
21 |
454 |
| 7.58e-206 |
26 |
462 |
28 |
463 |
| 7.58e-206 |
26 |
462 |
28 |
463 |
| 4.43e-157 |
26 |
459 |
33 |
466 |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 8.57e-12 |
140 |
361 |
119 |
324 |
Structure of N2152 from Neocallimastix frontalis [Neocallimastix frontalis] |
ACLA_097280-t26_1-p1 has no Swissprot hit.
This protein is predicted as SP
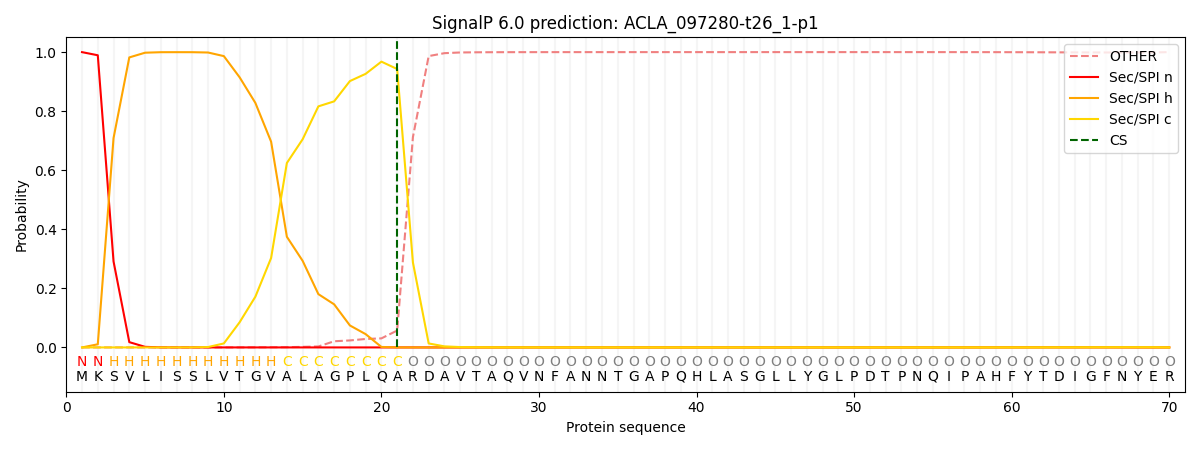
| Other |
SP_Sec_SPI |
CS Position |
| 0.001380 |
0.998578 |
CS pos: 21-22. Pr: 0.9427 |
There is no transmembrane helices in ACLA_097280-t26_1-p1.

