You are browsing environment: FUNGIDB
CAZyme Information: ACLA_078510-t26_1-p1
You are here: Home > Sequence: ACLA_078510-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus clavatus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus clavatus | |||||||||||
| CAZyme ID | ACLA_078510-t26_1-p1 | |||||||||||
| CAZy Family | GT109 | |||||||||||
| CAZyme Description | glycosyl hydrolase, putative | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 311649; End:314377 Strand: + | |||||||||||
Full Sequence Download help
| MGMGMGMGMG RAIVAISAVL LTARAQNIVN SPGDTAIIPG WSMQSSLRAP PNLSSLSLPG | 60 |
| EDVDLSSWYR VSSRATVMAG LVENGVYNDT TLFYSDNMET SIVDRAAFDA PWLYREEFTV | 120 |
| GKPGKGEHLF LKTHGITSKA DIYLNGALVA PAAVQQGSYG GHTYDITGIL RPGQNVLLVQ | 180 |
| ACPTNYLRDF AQGFVDWNPY PADNGTGVWR HVEVSKTGPV SISSPRVLTD FARPNGKPVT | 240 |
| VNVLVDVTNH DTQGVKGLLQ GTIQLDSLDI PISQTFSIKA HETKVVTINA KISSPEIWWP | 300 |
| GQWGSQPLYT LSINATVANA VSDVARPRRF GVRQVTSHLN AHNDTAFTVN DAPFLVLGGG | 360 |
| YAPDLFLRFD RVRVRKIFQY MLDIGLNTVR LEGNQEHPEL YDLADEMGLM VMAGWKCCDK | 420 |
| WEGWDYNNEA DGDKWTAKDY PVANASMIHE AAMMQTHPSM LAFLVGSDFW PNDRATKIYL | 480 |
| DALHRMDWPV PIIASASKRG YPQPLGPSGM KMDGPYDWVP PNYFYGDQMG AAFGFGSEQG | 540 |
| SGVGTPELPS LKKFLSPAEL ESLWKKPKQD QYHMSRYDSS FYNRALYNKA LFARYGKPGS | 600 |
| LDDYLRKAQM MDYETTRAEF EAFSVRQNAT RPATGVVYWM LNSAWPNLHW QLFDYYLSPA | 660 |
| GAYFGAKVGA RMEHVAYDYE AQAVYLVSHA LQSSGARVVV SVDLVDLQGQ SISHQEVTVD | 720 |
| SSPSASKVVA TATGIDQLKD VGFLRLRLQD VETGAVLSRN VYWVSAHNDA LDWDHSDLYY | 780 |
| TPVTKYANYQ ALGAMSPASI TASVKPLPAQ NGLNQVQVTL DDASTIPAFF VRLAVFDKDT | 840 |
| QEEITPVYWS DNYVTVFHGE QLTLTVAFAG DVSHSQIVLS GGNVGRTVLS PA | 892 |
Enzyme Prediction help
| EC | 3.2.1.165:3 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 105 | 716 | 1e-57 | 0.6170212765957447 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 408168 | Ig_GlcNase | 1.35e-36 | 779 | 884 | 2 | 104 | Exo-beta-D-glucosaminidase Ig-fold domain. This domain can be found in 2 glycoside hydrolase subfamily of beta-glucosaminidases (EC:3.2.1.165) such as CsxA, from Amycolatopsis orientalis that has exo-beta-D-glucosaminidase (exo-chitosanase) activity. It has an immunoglobulin-like topology. |
| 225789 | LacZ | 1.80e-20 | 108 | 663 | 61 | 623 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| 236657 | PRK10150 | 4.12e-13 | 63 | 411 | 30 | 352 | beta-D-glucuronidase; Provisional |
| 236673 | ebgA | 2.68e-12 | 113 | 413 | 112 | 396 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| 395572 | Glyco_hydro_2 | 3.59e-08 | 220 | 333 | 1 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ACO94980.1|GH2|3.2.1.165 | 0.0 | 28 | 890 | 23 | 883 |
| UDD56792.1|GH2 | 0.0 | 11 | 884 | 4 | 872 |
| AHJ61328.1|GH2|3.2.1.165 | 0.0 | 11 | 884 | 4 | 872 |
| QMW40806.1|GH2 | 0.0 | 11 | 884 | 4 | 872 |
| QRD83268.1|GH2 | 0.0 | 11 | 884 | 4 | 872 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2VZS_A | 1.88e-183 | 33 | 885 | 49 | 892 | Chitosan Product complex of Amycolatopsis orientalis exo-chitosanase CsxA [Amycolatopsis orientalis],2VZS_B Chitosan Product complex of Amycolatopsis orientalis exo-chitosanase CsxA [Amycolatopsis orientalis],2X05_A Inhibition of the exo-beta-D-glucosaminidase CsxA by a glucosamine- configured castanospermine and an amino-australine analogue [Amycolatopsis orientalis],2X05_B Inhibition of the exo-beta-D-glucosaminidase CsxA by a glucosamine- configured castanospermine and an amino-australine analogue [Amycolatopsis orientalis],2X09_A Inhibition of the exo-beta-D-glucosaminidase CsxA by a glucosamine- configured castanospermine and an amino-australine analogue [Amycolatopsis orientalis],2X09_B Inhibition of the exo-beta-D-glucosaminidase CsxA by a glucosamine- configured castanospermine and an amino-australine analogue [Amycolatopsis orientalis] |
| 2VZO_A | 7.37e-183 | 33 | 885 | 49 | 892 | Crystal structure of Amycolatopsis orientalis exo-chitosanase CsxA [Amycolatopsis orientalis],2VZO_B Crystal structure of Amycolatopsis orientalis exo-chitosanase CsxA [Amycolatopsis orientalis] |
| 2VZT_A | 1.46e-182 | 33 | 885 | 49 | 892 | Complex of Amycolatopsis orientalis exo-chitosanase CsxA E541A with PNP-beta-D-glucosamine [Amycolatopsis orientalis],2VZT_B Complex of Amycolatopsis orientalis exo-chitosanase CsxA E541A with PNP-beta-D-glucosamine [Amycolatopsis orientalis],2VZV_A Substrate Complex of Amycolatopsis orientalis exo-chitosanase CsxA E541A with chitosan [Amycolatopsis orientalis],2VZV_B Substrate Complex of Amycolatopsis orientalis exo-chitosanase CsxA E541A with chitosan [Amycolatopsis orientalis] |
| 2VZU_A | 4.45e-181 | 33 | 885 | 49 | 892 | Complex of Amycolatopsis orientalis exo-chitosanase CsxA D469A with PNP-beta-D-glucosamine [Amycolatopsis orientalis],2VZU_B Complex of Amycolatopsis orientalis exo-chitosanase CsxA D469A with PNP-beta-D-glucosamine [Amycolatopsis orientalis] |
| 5N6U_A | 1.84e-17 | 35 | 667 | 12 | 650 | Crystal structure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12],5N6U_B Crystal structure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12],5N6U_C Crystal structure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12],5N6U_D Crystal structure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| sp|D4AUH1|EBDG_ARTBC | 0.0 | 15 | 889 | 9 | 882 | Exo-beta-D-glucosaminidase ARB_07888 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_07888 PE=1 SV=1 |
| sp|Q4R1C4|EBDG_HYPJE | 7.07e-296 | 15 | 889 | 5 | 890 | Exo-beta-D-glucosaminidase OS=Hypocrea jecorina OX=51453 GN=gls93 PE=1 SV=1 |
| sp|C0LRA7|EBDG_HYPVI | 5.45e-287 | 15 | 889 | 5 | 888 | Exo-beta-D-glucosaminidase OS=Hypocrea virens OX=29875 GN=gls1 PE=2 SV=1 |
| sp|Q82NR8|EBDG_STRAW | 2.73e-200 | 24 | 892 | 40 | 904 | Exo-beta-D-glucosaminidase OS=Streptomyces avermitilis (strain ATCC 31267 / DSM 46492 / JCM 5070 / NBRC 14893 / NCIMB 12804 / NRRL 8165 / MA-4680) OX=227882 GN=csxA PE=1 SV=1 |
| sp|Q56F26|EBDG_AMYOR | 9.66e-183 | 33 | 885 | 49 | 892 | Exo-beta-D-glucosaminidase OS=Amycolatopsis orientalis OX=31958 GN=csxA PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
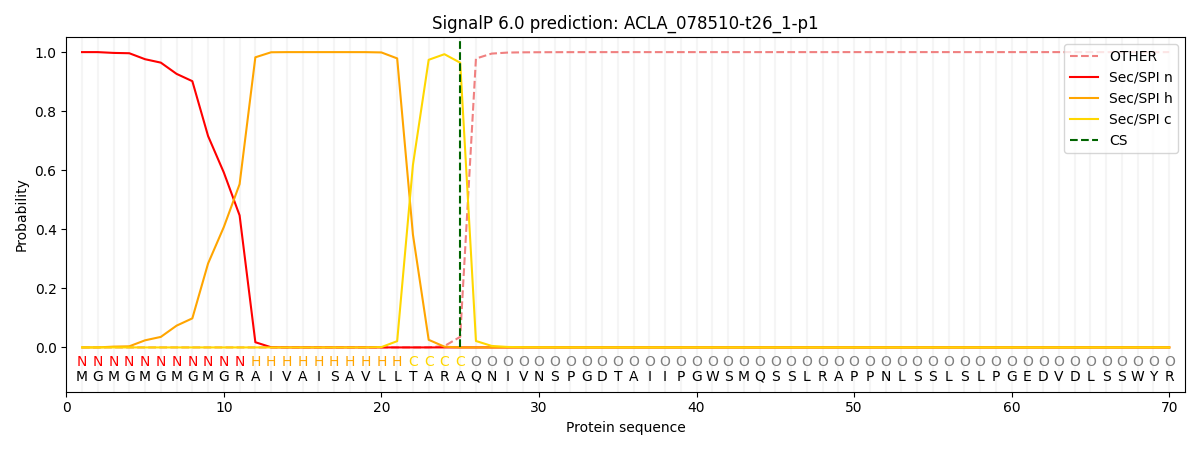
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000751 | 0.999220 | CS pos: 25-26. Pr: 0.9640 |
