You are browsing environment: FUNGIDB
CAZyme Information: ACLA_071530-t26_1-p1
You are here: Home > Sequence: ACLA_071530-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus clavatus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus clavatus | |||||||||||
| CAZyme ID | ACLA_071530-t26_1-p1 | |||||||||||
| CAZy Family | GH7 | |||||||||||
| CAZyme Description | cellulose hydrolase, putative | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.8:3 | 3.2.1.-:3 | 3.2.1.4:10 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH30 | 18 | 447 | 3.7e-185 | 0.9909706546275395 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 227807 | XynC | 6.16e-22 | 21 | 397 | 38 | 400 | O-Glycosyl hydrolase [Cell wall/membrane/envelope biogenesis]. |
| 197593 | fCBD | 2.91e-14 | 478 | 511 | 1 | 34 | Fungal-type cellulose-binding domain. Small four-cysteine cellulose-binding domain of fungi |
| 395595 | CBM_1 | 4.33e-14 | 479 | 507 | 1 | 29 | Fungal cellulose binding domain. |
| 396577 | Glyco_hydro_59 | 5.15e-13 | 31 | 362 | 2 | 283 | Glycosyl hydrolase family 59. |
| 405300 | Glyco_hydr_30_2 | 5.71e-12 | 21 | 322 | 6 | 353 | O-Glycosyl hydrolase family 30. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.45e-163 | 26 | 511 | 31 | 527 | |
| 3.29e-157 | 4 | 511 | 7 | 533 | |
| 5.75e-153 | 26 | 448 | 35 | 467 | |
| 9.99e-153 | 26 | 448 | 61 | 493 | |
| 6.68e-152 | 26 | 511 | 32 | 522 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.33e-140 | 26 | 448 | 12 | 447 | Chain A, GH30 family xylanase [Thermothelomyces thermophilus ATCC 42464],7O0E_G Chain G, GH30 family xylanase [Thermothelomyces thermophilus ATCC 42464] |
|
| 2.09e-139 | 26 | 449 | 32 | 454 | Chain A, GH30 Xylanase C [Talaromyces cellulolyticus CF-2612],6M5Z_B Chain B, GH30 Xylanase C [Talaromyces cellulolyticus CF-2612] |
|
| 7.17e-139 | 26 | 448 | 19 | 454 | Chain AAA, GH30 family xylanase [Thermothelomyces thermophilus ATCC 42464] |
|
| 3.24e-121 | 27 | 402 | 32 | 421 | Chain A, GH30 Xylanase B [Talaromyces cellulolyticus CF-2612],6IUJ_B Chain B, GH30 Xylanase B [Talaromyces cellulolyticus CF-2612] |
|
| 2.86e-120 | 27 | 402 | 103 | 492 | Crystal structure of GH30 xylanase B from Talaromyces cellulolyticus expressed by Pichia pastoris [Saccharomyces uvarum],6KRN_A Crystal structure of GH30 xylanase B from Talaromyces cellulolyticus expressed by Pichia pastoris in complex with aldotriuronic acid [Saccharomyces uvarum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.86e-140 | 26 | 448 | 36 | 471 | GH30 family xylanase OS=Myceliophthora thermophila (strain ATCC 42464 / BCRC 31852 / DSM 1799) OX=573729 GN=Xyn30A PE=1 SV=1 |
|
| 6.21e-10 | 28 | 402 | 71 | 463 | Endo-1,6-beta-D-glucanase neg1 OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=neg1 PE=1 SV=1 |
|
| 6.79e-10 | 480 | 511 | 506 | 537 | Exoglucanase 1 OS=Penicillium janthinellum OX=5079 GN=cbh1 PE=2 SV=1 |
|
| 4.18e-09 | 476 | 511 | 336 | 371 | Probable acetylxylan esterase A OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=axeA PE=3 SV=1 |
|
| 4.22e-09 | 480 | 511 | 433 | 464 | Endoglucanase 7a OS=Myceliophthora thermophila (strain ATCC 42464 / BCRC 31852 / DSM 1799) OX=573729 GN=eg7A PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
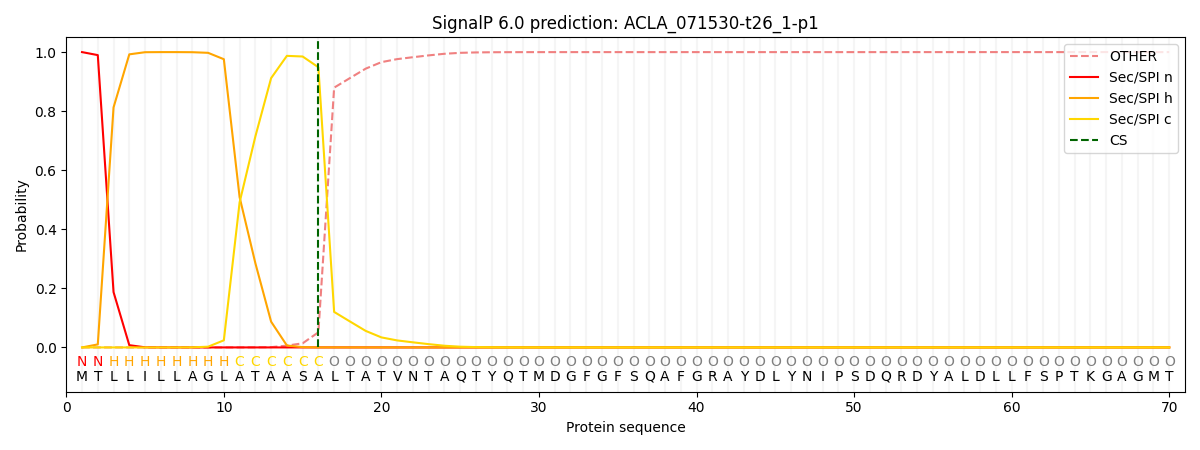
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000260 | 0.999717 | CS pos: 16-17. Pr: 0.9486 |
