You are browsing environment: FUNGIDB
CAZyme Information: ACLA_062560-t26_1-p1
You are here: Home > Sequence: ACLA_062560-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus clavatus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus clavatus | |||||||||||
| CAZyme ID | ACLA_062560-t26_1-p1 | |||||||||||
| CAZy Family | GH47 | |||||||||||
| CAZyme Description | cellobiohydrolase, putative | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.91:55 | 3.2.1.4:4 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH6 | 132 | 425 | 2.2e-91 | 0.9387755102040817 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396075 | Glyco_hydro_6 | 8.39e-163 | 134 | 428 | 1 | 293 | Glycosyl hydrolases family 6. |
| 227616 | CelA1 | 8.01e-55 | 97 | 463 | 12 | 442 | Cellulase/cellobiase CelA1 [Carbohydrate transport and metabolism]. |
| 395595 | CBM_1 | 2.96e-10 | 23 | 51 | 1 | 29 | Fungal cellulose binding domain. |
| 197593 | fCBD | 2.00e-09 | 24 | 55 | 3 | 34 | Fungal-type cellulose-binding domain. Small four-cysteine cellulose-binding domain of fungi |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 7.87e-251 | 1 | 464 | 1 | 454 | |
| 1.38e-237 | 9 | 464 | 8 | 464 | |
| 1.38e-237 | 9 | 464 | 8 | 464 | |
| 1.38e-237 | 9 | 464 | 8 | 464 | |
| 1.38e-237 | 9 | 464 | 8 | 464 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.10e-180 | 102 | 464 | 1 | 364 | Chain A, Cellobiohydrolase Family 6 [Thermochaetoides thermophila] |
|
| 2.98e-177 | 102 | 464 | 1 | 362 | Chain A, CELLOBIOHYDROLASE CEL6A (FORMERLY CALLED CBH II) [Trichoderma reesei],1QK0_B Chain B, CELLOBIOHYDROLASE CEL6A (FORMERLY CALLED CBH II) [Trichoderma reesei],1QK2_A Chain A, CELLOBIOHYDROLASE CEL6A (FORMERLY CALLED CBH II) [Trichoderma reesei],1QK2_B Chain B, CELLOBIOHYDROLASE CEL6A (FORMERLY CALLED CBH II) [Trichoderma reesei] |
|
| 3.20e-177 | 102 | 464 | 3 | 364 | Chain A, CELLOBIOHYDROLASE II CORE PROTEIN [Trichoderma reesei] |
|
| 1.30e-176 | 102 | 464 | 3 | 364 | Chain A, CELLOBIOHYDROLASE II [Trichoderma reesei],1CB2_B Chain B, CELLOBIOHYDROLASE II [Trichoderma reesei],1QJW_A Chain A, CELLOBIOHYDROLASE CEL6A (FORMERLY CALLED CBH II) [Trichoderma reesei],1QJW_B Chain B, CELLOBIOHYDROLASE CEL6A (FORMERLY CALLED CBH II) [Trichoderma reesei] |
|
| 3.57e-176 | 107 | 464 | 1 | 357 | Crystal structure of a fungal chimeric cellobiohydrolase Cel6A [Humicola insolens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 0.0 | 1 | 464 | 1 | 464 | Probable 1,4-beta-D-glucan cellobiohydrolase C OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=cbhC PE=3 SV=1 |
|
| 5.98e-252 | 1 | 464 | 1 | 450 | Probable 1,4-beta-D-glucan cellobiohydrolase C OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=cbhC PE=3 SV=1 |
|
| 1.40e-251 | 1 | 464 | 1 | 454 | Probable 1,4-beta-D-glucan cellobiohydrolase C OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=cbhC PE=3 SV=1 |
|
| 1.40e-251 | 1 | 464 | 1 | 454 | Probable 1,4-beta-D-glucan cellobiohydrolase C OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=cbhC PE=3 SV=1 |
|
| 7.40e-248 | 7 | 464 | 6 | 468 | Probable 1,4-beta-D-glucan cellobiohydrolase C OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=cbhC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
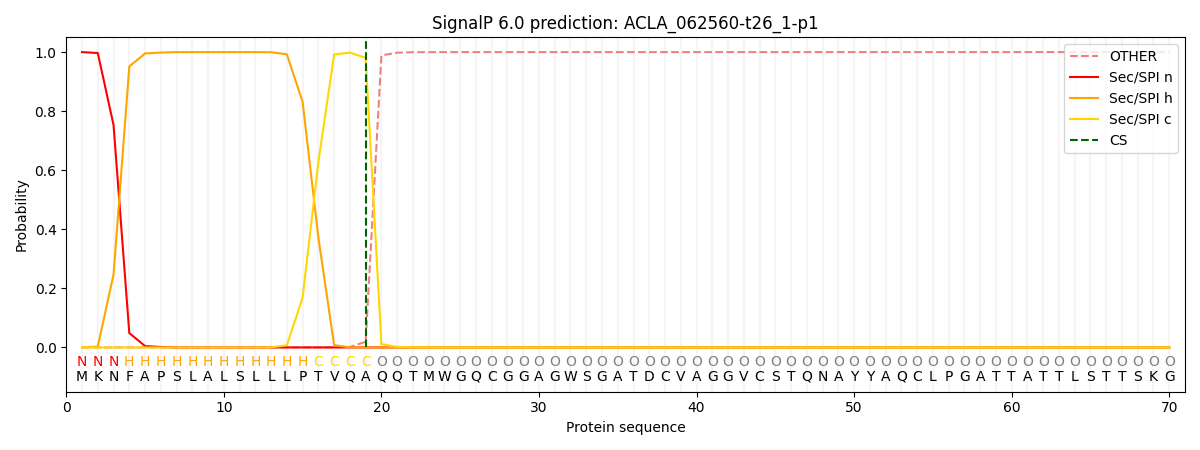
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000246 | 0.999734 | CS pos: 19-20. Pr: 0.9807 |
