You are browsing environment: FUNGIDB
CAZyme Information: ACLA_035050-t26_1-p1
You are here: Home > Sequence: ACLA_035050-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus clavatus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus clavatus | |||||||||||
| CAZyme ID | ACLA_035050-t26_1-p1 | |||||||||||
| CAZy Family | GH13 | |||||||||||
| CAZyme Description | MFS transporter, putative | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.14:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH18 | 32 | 280 | 6.8e-23 | 0.6655405405405406 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 119356 | GH18_hevamine_XipI_class_III | 5.76e-107 | 33 | 313 | 3 | 267 | This conserved domain family includes xylanase inhibitor Xip-I, and the class III plant chitinases such as hevamine, concanavalin B, and PPL2, all of which have a glycosyl hydrolase family 18 (GH18) domain. Hevamine is a class III endochitinase that hydrolyzes the linear polysaccharide chains of chitin and peptidoglycan and is important for defense against pathogenic bacteria and fungi. PPL2 (Parkia platycephala lectin 2) is a class III chitinase from Parkia platycephala seeds that hydrolyzes beta(1-4) glycosidic bonds linking 2-acetoamido-2-deoxy-beta-D-glucopyranose units in chitin. |
| 340881 | MFS_Tpo1_MDR_like | 6.65e-49 | 471 | 1052 | 6 | 376 | Yeast Polyamine transporter 1 (Tpo1) and similar multidrug resistance (MDR) transporters of the Major Facilitator Superfamily. This family is composed of fungal multidrug resistance (MDR) transporters including several proteins from Saccharomyces cerevisiae such as polyamine transporters 1-4 (Tpo1-4), quinidine resistance proteins 1-3 (Qdr1-3), dityrosine transporter 1 (Dtr1), fluconazole resistance protein 1 (Flr1), and protein HOL1. MDR transporters are drug/H+ antiporters (DHA) that mediate the efflux of a variety of drugs and toxic compounds, and confer resistance to these compounds. For example, Flr1 confers resistance to the azole derivative fluconazole while Tpo1 confers resistance and adaptation to quinidine and ketoconazole. The polyamine transporters are involved in the detoxification of excess polyamines in the cytoplasm. Tpo1-like MDR transporters belong to the Major Facilitator Superfamily (MFS) of membrane transport proteins, which are thought to function through a single substrate binding site, alternating-access mechanism involving a rocker-switch type of movement. |
| 273318 | 2_A_01_02 | 8.61e-26 | 509 | 645 | 1 | 141 | Multidrug resistance protein. |
| 340878 | MFS_MdfA_MDR_like | 1.50e-23 | 482 | 645 | 18 | 177 | Multidrug transporter MdfA and similar multidrug resistance (MDR) transporters of the Major Facilitator Superfamily. This family is composed of bacterial multidrug resistance (MDR) transporters including several proteins from Escherichia coli such as MdfA (also called chloramphenicol resistance pump Cmr), EmrD, MdtM, MdtL, bicyclomycin resistance protein (also called sulfonamide resistance protein), and the uncharacterized inner membrane transport protein YdhC. EmrD is a proton-dependent secondary transporter, first identified as an efflux pump for uncouplers of oxidative phosphorylation. It expels a range of drug molecules and amphipathic compounds across the inner membrane of E. coli. Similarly, MdfA is a secondary multidrug transporter that exports a broad spectrum of structurally and electrically dissimilar toxic compounds. These MDR transporters are drug/H+ antiporters (DHA) belonging to the Major Facilitator Superfamily (MFS) of membrane transport proteins, which are thought to function through a single substrate binding site, alternating-access mechanism involving a rocker-switch type of movement. |
| 369468 | MFS_1 | 8.33e-23 | 469 | 645 | 1 | 175 | Major Facilitator Superfamily. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 8.62e-163 | 1 | 314 | 1 | 313 | |
| 8.74e-118 | 10 | 312 | 7 | 313 | |
| 4.71e-117 | 10 | 312 | 7 | 313 | |
| 2.71e-116 | 25 | 312 | 24 | 315 | |
| 1.36e-113 | 8 | 312 | 7 | 334 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.61e-57 | 26 | 312 | 1 | 267 | ScCTS1_apo crystal structure [Saccharomyces cerevisiae],2UY3_A ScCTS1_8-chlorotheophylline crystal structure [Saccharomyces cerevisiae],2UY4_A ScCTS1_acetazolamide crystal structure [Saccharomyces cerevisiae],2UY5_A ScCTS1_kinetin crystal structure [Saccharomyces cerevisiae],4TXE_A ScCTS1 in complex with compound 5 [Saccharomyces cerevisiae] |
|
| 6.38e-46 | 31 | 313 | 1 | 288 | A. fumigatus chitinase A1 phenyl-methylguanylurea complex [Aspergillus fumigatus],2XVN_B A. fumigatus chitinase A1 phenyl-methylguanylurea complex [Aspergillus fumigatus],2XVN_C A. fumigatus chitinase A1 phenyl-methylguanylurea complex [Aspergillus fumigatus] |
|
| 6.55e-46 | 31 | 313 | 2 | 289 | ChiA1 from Aspergillus fumigatus in complex with acetazolamide [Aspergillus fumigatus A1163],2XTK_B ChiA1 from Aspergillus fumigatus in complex with acetazolamide [Aspergillus fumigatus A1163],2XUC_A Natural product-guided discovery of a fungal chitinase inhibitor [Aspergillus fumigatus],2XUC_B Natural product-guided discovery of a fungal chitinase inhibitor [Aspergillus fumigatus],2XUC_C Natural product-guided discovery of a fungal chitinase inhibitor [Aspergillus fumigatus],2XVP_A ChiA1 from Aspergillus fumigatus, apostructure [Aspergillus fumigatus A1163],2XVP_B ChiA1 from Aspergillus fumigatus, apostructure [Aspergillus fumigatus A1163] |
|
| 6.55e-46 | 31 | 313 | 2 | 289 | AfChiA1 in complex with compound 1 [Aspergillus fumigatus A1163],4TX6_B AfChiA1 in complex with compound 1 [Aspergillus fumigatus A1163] |
|
| 5.76e-36 | 33 | 310 | 3 | 254 | cDNA cloning and 1.75A crystal structure determination of PPL2, a novel chimerolectin from Parkia platycephala seeds exhibiting endochitinolytic activity [Parkia platycephala] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.28e-98 | 10 | 312 | 6 | 295 | Class III chitinase ARB_03514 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_03514 PE=1 SV=2 |
|
| 2.37e-82 | 10 | 314 | 3 | 306 | Endochitinase 33 OS=Trichoderma harzianum OX=5544 GN=chit33 PE=1 SV=1 |
|
| 1.02e-57 | 27 | 312 | 25 | 295 | Chitinase 2 OS=Rhizopus oligosporus OX=4847 GN=CHI2 PE=1 SV=1 |
|
| 8.39e-57 | 27 | 312 | 25 | 295 | Chitinase 1 OS=Rhizopus oligosporus OX=4847 GN=CHI1 PE=1 SV=1 |
|
| 1.67e-55 | 25 | 345 | 17 | 318 | Chitinase 3 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=CHT3 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000294 | 0.999675 | CS pos: 23-24. Pr: 0.9770 |
TMHMM Annotations download full data without filtering help
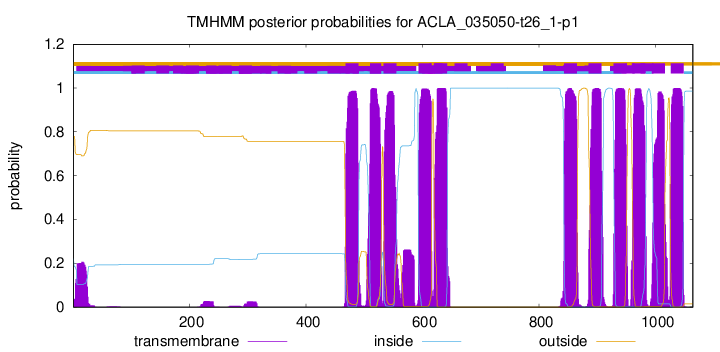
| Start | End |
|---|---|
| 468 | 490 |
| 510 | 529 |
| 533 | 555 |
| 593 | 615 |
| 620 | 642 |
| 843 | 865 |
| 885 | 907 |
| 928 | 950 |
| 960 | 982 |
| 994 | 1016 |
| 1026 | 1048 |
