You are browsing environment: FUNGIDB
CAZyme Information: ACLA_017480-t26_1-p1
You are here: Home > Sequence: ACLA_017480-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus clavatus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus clavatus | |||||||||||
| CAZyme ID | ACLA_017480-t26_1-p1 | |||||||||||
| CAZy Family | AA9 | |||||||||||
| CAZyme Description | glycosyl hydrolase family 62 protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE1 | 45 | 173 | 9e-24 | 0.5814977973568282 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 226040 | LpqC | 2.13e-21 | 44 | 269 | 46 | 304 | Poly(3-hydroxybutyrate) depolymerase [Secondary metabolites biosynthesis, transport and catabolism]. |
| 226584 | COG4099 | 1.38e-06 | 47 | 203 | 176 | 337 | Predicted peptidase [General function prediction only]. |
| 223489 | DLH | 7.12e-06 | 120 | 219 | 96 | 196 | Dienelactone hydrolase [Secondary metabolites biosynthesis, transport and catabolism]. |
| 395613 | Esterase | 1.27e-04 | 44 | 169 | 5 | 143 | Putative esterase. This family contains Esterase D. However it is not clear if all members of the family have the same function. This family is related to the pfam00135 family. |
| 223477 | YpfH | 0.001 | 113 | 207 | 76 | 172 | Predicted esterase [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.21e-87 | 22 | 269 | 90 | 352 | |
| 4.18e-85 | 28 | 269 | 38 | 285 | |
| 2.68e-80 | 24 | 269 | 55 | 321 | |
| 3.94e-80 | 24 | 269 | 18 | 279 | |
| 7.70e-79 | 19 | 269 | 31 | 296 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.72e-203 | 1 | 272 | 1 | 272 | Probable feruloyl esterase C OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=faeC-2 PE=3 SV=1 |
|
| 5.24e-160 | 1 | 272 | 1 | 272 | Probable feruloyl esterase C OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=faeC PE=3 SV=1 |
|
| 5.24e-160 | 1 | 272 | 1 | 272 | Probable feruloyl esterase C OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=faeC PE=3 SV=1 |
|
| 3.52e-158 | 1 | 272 | 1 | 272 | Probable feruloyl esterase C OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=faeC PE=3 SV=1 |
|
| 8.78e-152 | 1 | 272 | 1 | 272 | Probable feruloyl esterase C OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=faeC-1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
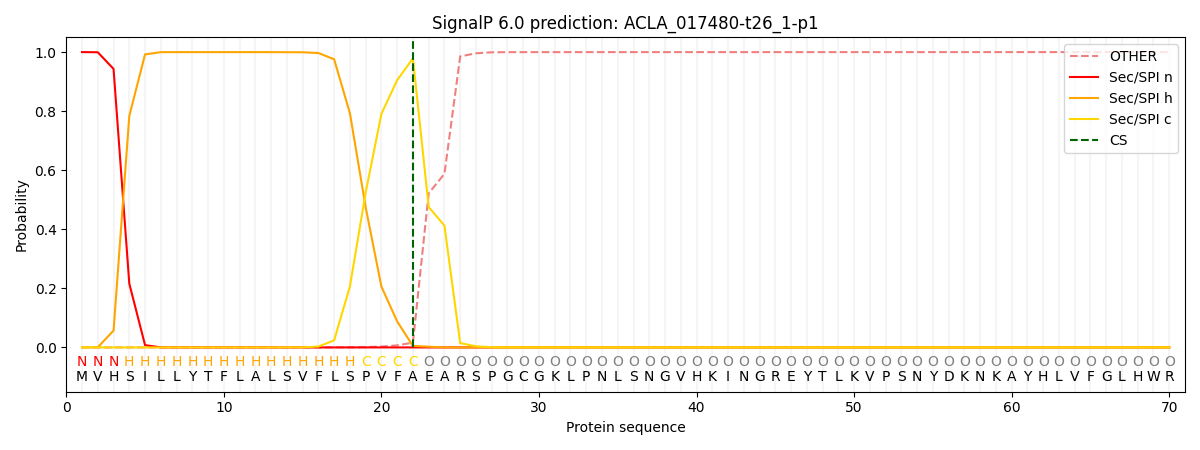
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000198 | 0.999762 | CS pos: 22-23. Pr: 0.9787 |
