You are browsing environment: FUNGIDB
CAZyme Information: ACLA_008190-t26_1-p1
You are here: Home > Sequence: ACLA_008190-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus clavatus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus clavatus | |||||||||||
| CAZyme ID | ACLA_008190-t26_1-p1 | |||||||||||
| CAZy Family | AA7 | |||||||||||
| CAZyme Description | chitin binding domain protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 1.14.99.55:5 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA13 | 19 | 247 | 1.1e-137 | 0.9956896551724138 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 412056 | AA13_LPMO-like | 1.93e-160 | 19 | 248 | 1 | 233 | AA13 lytic polysaccharide monooxygenase, and similar proteins. This family contains starch-degrading (also called starch-active) lytic polysaccharide monooxygenase (LPMO), a representative of the new CAZy AA13 family and classified as an auxiliary activity enzyme. This enzyme acts on alpha-linked glycosidic bonds and displays a binding surface that is quite different from those of LPMOs acting on beta-linked glycosidic bonds, indicating that the AA13 family proteins interact with their substrate in a distinct fashion. The active site contains an amino-terminal histidine-ligated mononuclear copper. This enzyme generates aldonic acid-terminated malto-oligosaccharides from retrograded starch and significantly boosts the conversion of this recalcitrant substrate to maltose by beta-amylase. |
| 397269 | LPMO_10 | 0.006 | 19 | 120 | 1 | 117 | Lytic polysaccharide mono-oxygenase, cellulose-degrading. This domain is found associated with a wide variety of cellulose binding domains. This is a family of two very closely related proteins that together act as both a C1- and a C4-oxidising lytic polysaccharide mono-oxygenase, degrading cellulose. This domain is also found in baculoviral spheroidins and spindolins, protein of unknown function. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.62e-154 | 1 | 247 | 1 | 250 | |
| 1.62e-154 | 1 | 247 | 1 | 250 | |
| 1.62e-154 | 1 | 247 | 1 | 250 | |
| 1.62e-154 | 1 | 247 | 1 | 250 | |
| 1.48e-152 | 4 | 247 | 32 | 278 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.86e-147 | 20 | 247 | 2 | 232 | AA13 Lytic polysaccharide monooxygenase from Aspergillus oryzae [Aspergillus oryzae RIB40],5LSV_A X-ray crystal structure of AA13 LPMO [Aspergillus oryzae RIB40],5T7J_A X-ray crystal structure of AA13 LPMO [Aspergillus oryzae RIB40],5T7K_A X-ray crystal structure of AA13 LPMO [Aspergillus oryzae RIB40],5T7N_A X-ray crystal structure of AA13 LPMO [Aspergillus oryzae RIB40],6TBQ_A AA13 Lytic polysaccharide monooxygenase from Aspergillus oryzae partially in Cu(II) state [Aspergillus oryzae RIB40],6TBR_A Glycosylated AA13 Lytic polysaccharide monooxygenase from Aspergillus oryzae in P1 space group [Aspergillus oryzae RIB40],6TBR_B Glycosylated AA13 Lytic polysaccharide monooxygenase from Aspergillus oryzae in P1 space group [Aspergillus oryzae RIB40],6TC4_A AA13 Lytic polysaccharide monooxygenase from Aspergillus oryzae measured with SSX [Aspergillus oryzae RIB40] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
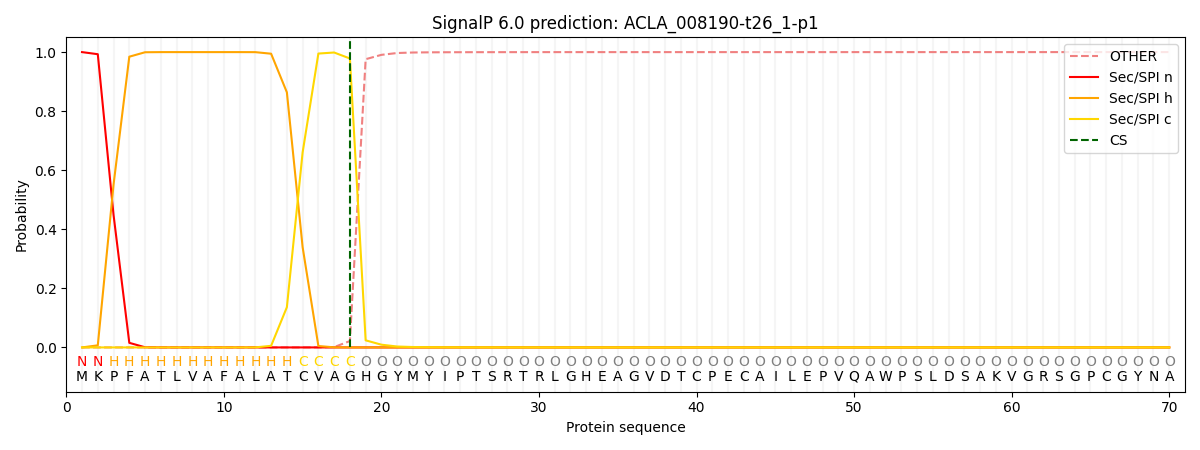
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000256 | 0.999715 | CS pos: 18-19. Pr: 0.9784 |
