You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004726_00837
You are here: Home > Sequence: MGYG000004726_00837
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
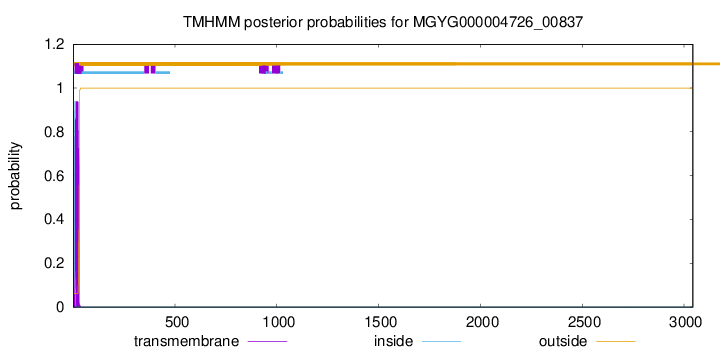
TMHMM annotations
Basic Information help
| Species | Pseudoflavonifractor sp014287675 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Oscillospiraceae; Pseudoflavonifractor; Pseudoflavonifractor sp014287675 | |||||||||||
| CAZyme ID | MGYG000004726_00837 | |||||||||||
| CAZy Family | GH101 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 148001; End: 157135 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH101 | 869 | 1409 | 4.4e-85 | 0.7934936350777935 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd14244 | GH_101_like | 5.84e-57 | 1132 | 1406 | 4 | 298 | Endo-a-N-acetylgalactosaminidase and related glcyosyl hydrolases. This family contains the enzymatically active domain of cell surface proteins that specifically cleave Gal-beta-1,3-GalNAc-alpha-Ser/Thr (T-antigen, galacto-N-biose), the core 1 type O-linked glycan common to mucin glycoproteins (EC:3.2.1.97). It has been classified as glycosyl hydrolase family 101 in the Cazy resource. Virulence of pathogenic organisms such as the Gram-positive Streptococcus pneumoniae and other commensal human bacteria is largely determined by their ability to degrade host glycoproteins and to metabolize the resultant carbohydrates. |
| pfam12905 | Glyco_hydro_101 | 4.85e-55 | 1116 | 1380 | 1 | 270 | Endo-alpha-N-acetylgalactosaminidase. Virulence of pathogenic organisms such as the Gram-positive Streptococcus pneumoniae is largely determined by the ability to degrade host glycoproteins and to metabolize the resultant carbohydrates. This family is the enzymatic region, EC:3.2.1.97, of the cell surface proteins that specifically cleave Gal-beta-1,3-GalNAc-alpha-Ser/Thr (T-antigen, galacto-N-biose), the core 1 type O-linked glycan common to mucin glycoproteins. This reaction is exemplified by the S. pneumoniae protein Endo-alpha-N-acetylgalactosaminidase, where Asp764 is the catalytic nucleophile-base and Glu796 the catalytic proton donor. |
| pfam18080 | Gal_mutarotas_3 | 1.87e-35 | 864 | 1115 | 1 | 243 | Galactose mutarotase-like fold domain. This domain is found in endo-alpha-N-acetylgalactosaminidase present in Streptococcus pneumoniae. Endo-alpha-N-acetylgalactosaminidase is a cell surface-anchored glycoside hydrolase involved in the breakdown of mucin type O-linked glycans. The domain, known as domain 2, exhibits strong structural similarlity to the galactose mutarotase-like fold but lacks the active site residues. Domains, found in a number of glycoside hydrolases, structurally similar to domain 2 confer stability to the multidomain architectures. |
| pfam13385 | Laminin_G_3 | 1.63e-26 | 103 | 260 | 1 | 151 | Concanavalin A-like lectin/glucanases superfamily. This domain belongs to the Concanavalin A-like lectin/glucanases superfamily. |
| pfam13385 | Laminin_G_3 | 7.18e-25 | 2060 | 2215 | 1 | 151 | Concanavalin A-like lectin/glucanases superfamily. This domain belongs to the Concanavalin A-like lectin/glucanases superfamily. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ATD58436.1 | 3.50e-152 | 372 | 1813 | 237 | 1608 |
| ATD54117.1 | 3.50e-152 | 372 | 1813 | 237 | 1608 |
| QBJ76357.1 | 3.50e-152 | 372 | 1813 | 237 | 1608 |
| SLK22612.1 | 3.50e-152 | 372 | 1813 | 237 | 1608 |
| BBK22105.1 | 1.53e-133 | 372 | 1827 | 28 | 1471 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6QFO_A | 2.48e-91 | 854 | 1823 | 2 | 1091 | EngBFDARPin Fusion 9b 3G124 [Bifidobacterium longum] |
| 6QEP_A | 5.32e-90 | 854 | 1823 | 2 | 1091 | EngBFDARPin Fusion 4b H14 [Bifidobacterium longum] |
| 6QFK_A | 1.71e-89 | 854 | 1823 | 2 | 1091 | EngBFDARPin Fusion 4b G10 [Bifidobacterium longum] |
| 6QEV_B | 1.71e-89 | 854 | 1823 | 2 | 1091 | EngBFDARPin Fusion 4b B6 [Bifidobacterium longum] |
| 6SH9_B | 1.71e-89 | 854 | 1823 | 2 | 1091 | EngBFDARPin Fusion 4b D12 [Bifidobacterium longum subsp. longum JCM 1217] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A9WNA0 | 2.36e-71 | 860 | 1768 | 48 | 962 | Putative endo-alpha-N-acetylgalactosaminidase OS=Renibacterium salmoninarum (strain ATCC 33209 / DSM 20767 / JCM 11484 / NBRC 15589 / NCIMB 2235) OX=288705 GN=RSal33209_1326 PE=3 SV=2 |
| Q8DR60 | 6.18e-70 | 863 | 1768 | 333 | 1319 | Endo-alpha-N-acetylgalactosaminidase OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=spr0328 PE=1 SV=1 |
| Q2MGH6 | 3.15e-69 | 863 | 1768 | 333 | 1319 | Endo-alpha-N-acetylgalactosaminidase OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=SP_0368 PE=1 SV=1 |
| P38535 | 2.19e-11 | 2875 | 3042 | 901 | 1084 | Exoglucanase XynX OS=Acetivibrio thermocellus OX=1515 GN=xynX PE=3 SV=1 |
| P38536 | 5.74e-11 | 2863 | 3042 | 1663 | 1858 | Amylopullulanase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=amyB PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
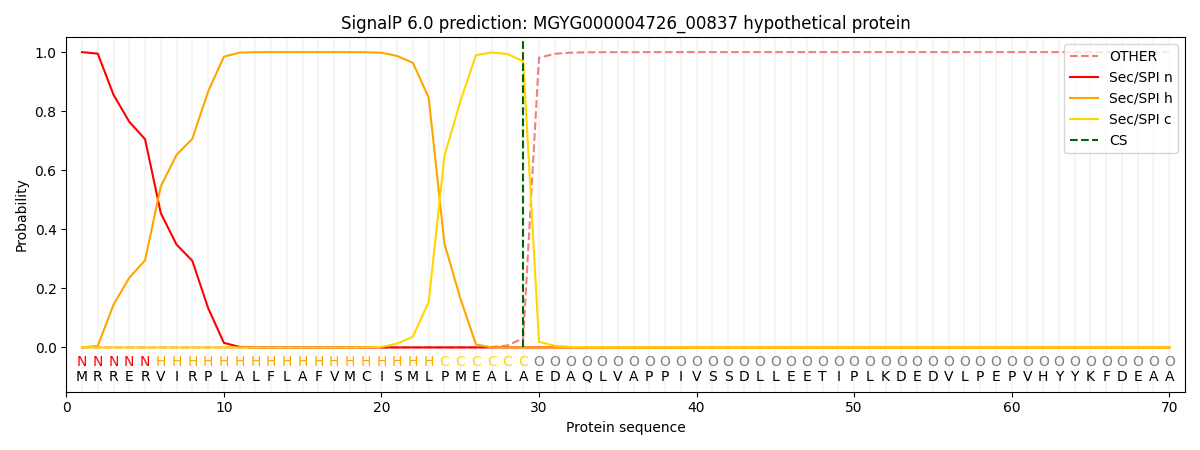
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000393 | 0.998646 | 0.000415 | 0.000185 | 0.000180 | 0.000159 |