You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004725_00806
You are here: Home > Sequence: MGYG000004725_00806
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
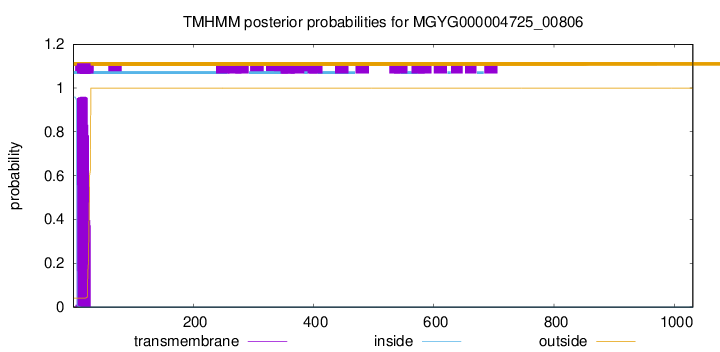
TMHMM annotations
Basic Information help
| Species | UBA9475 sp900554075 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Oscillospiraceae; UBA9475; UBA9475 sp900554075 | |||||||||||
| CAZyme ID | MGYG000004725_00806 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 77450; End: 80545 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 494 | 870 | 1.6e-41 | 0.9305555555555556 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4677 | PemB | 1.71e-22 | 477 | 852 | 60 | 375 | Pectin methylesterase and related acyl-CoA thioesterases [Carbohydrate transport and metabolism, Lipid transport and metabolism]. |
| pfam01095 | Pectinesterase | 2.67e-14 | 702 | 868 | 119 | 275 | Pectinesterase. |
| PLN02708 | PLN02708 | 3.11e-14 | 703 | 870 | 363 | 528 | Probable pectinesterase/pectinesterase inhibitor |
| PLN03043 | PLN03043 | 8.28e-14 | 681 | 867 | 319 | 500 | Probable pectinesterase/pectinesterase inhibitor; Provisional |
| PLN02773 | PLN02773 | 8.66e-14 | 687 | 888 | 121 | 305 | pectinesterase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AFK85374.1 | 1.36e-221 | 30 | 894 | 752 | 1724 |
| ADL35206.1 | 2.12e-197 | 28 | 914 | 132 | 1142 |
| AOW17789.1 | 7.41e-175 | 31 | 882 | 572 | 1507 |
| ACU08321.1 | 1.25e-169 | 33 | 963 | 23 | 1056 |
| QQY82325.1 | 5.64e-161 | 31 | 967 | 161 | 1185 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5C1C_A | 1.14e-12 | 684 | 850 | 112 | 260 | CrystalStructure of the Pectin Methylesterase from Aspergillus niger in Deglycosylated Form [Aspergillus niger ATCC 1015] |
| 5C1E_A | 1.14e-12 | 684 | 850 | 112 | 260 | CrystalStructure of the Pectin Methylesterase from Aspergillus niger in Penultimately Deglycosylated Form (N-acetylglucosamine Stub at Asn84) [Aspergillus niger ATCC 1015] |
| 3UW0_A | 2.10e-11 | 620 | 842 | 114 | 309 | Pectinmethylesterase from Yersinia enterocolitica [Yersinia enterocolitica subsp. enterocolitica 8081] |
| 2NSP_A | 1.00e-10 | 611 | 820 | 79 | 273 | ChainA, Pectinesterase A [Dickeya dadantii 3937],2NSP_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NST_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NST_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT6_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT6_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT9_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT9_B Chain B, Pectinesterase A [Dickeya dadantii 3937] |
| 1XG2_A | 2.59e-07 | 676 | 867 | 96 | 277 | ChainA, Pectinesterase 1 [Solanum lycopersicum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q4WBT5 | 4.40e-13 | 684 | 894 | 137 | 319 | Probable pectinesterase A OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=pmeA PE=3 SV=1 |
| B0Y9F9 | 4.40e-13 | 684 | 894 | 137 | 319 | Probable pectinesterase A OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=pmeA PE=3 SV=1 |
| Q8L7Q7 | 7.59e-13 | 676 | 870 | 384 | 577 | Probable pectinesterase/pectinesterase inhibitor 64 OS=Arabidopsis thaliana OX=3702 GN=PME64 PE=2 SV=2 |
| A1DBT4 | 1.42e-12 | 684 | 850 | 137 | 285 | Probable pectinesterase A OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=pmeA PE=3 SV=1 |
| A2QK82 | 8.42e-12 | 684 | 850 | 140 | 288 | Probable pectinesterase A OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=pmeA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
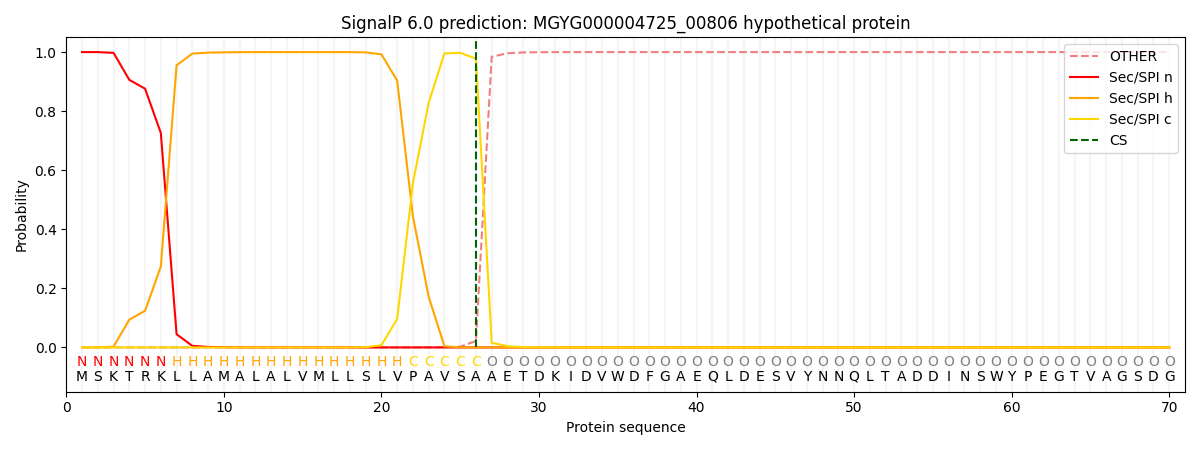
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000191 | 0.999158 | 0.000176 | 0.000172 | 0.000155 | 0.000144 |