You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004622_00043
You are here: Home > Sequence: MGYG000004622_00043
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900316565 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900316565 | |||||||||||
| CAZyme ID | MGYG000004622_00043 | |||||||||||
| CAZy Family | GH26 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 52658; End: 55003 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 469 | 753 | 4.4e-99 | 0.9818840579710145 |
| GH26 | 95 | 422 | 3.3e-87 | 0.9933993399339934 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 5.10e-51 | 468 | 752 | 15 | 266 | Cellulase (glycosyl hydrolase family 5). |
| pfam02156 | Glyco_hydro_26 | 2.31e-40 | 95 | 422 | 2 | 311 | Glycosyl hydrolase family 26. |
| COG2730 | BglC | 3.26e-18 | 440 | 713 | 50 | 323 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| COG4124 | ManB2 | 5.46e-12 | 222 | 347 | 154 | 263 | Beta-mannanase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNT65320.1 | 0.0 | 1 | 781 | 1 | 786 |
| AGH13958.1 | 0.0 | 1 | 780 | 1 | 775 |
| AHG56239.1 | 0.0 | 2 | 780 | 150 | 923 |
| AAC97596.1 | 0.0 | 2 | 780 | 150 | 923 |
| BCS86234.1 | 0.0 | 1 | 781 | 1 | 776 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6D2W_A | 0.0 | 1 | 780 | 1 | 775 | Crystalstructure of Prevotella bryantii endo-beta-mannanase/endo-beta-glucanase PbGH26A-GH5A [Prevotella bryantii B14],6D2W_B Crystal structure of Prevotella bryantii endo-beta-mannanase/endo-beta-glucanase PbGH26A-GH5A [Prevotella bryantii B14] |
| 6D2X_A | 3.94e-164 | 91 | 426 | 2 | 337 | Crystalstructure of the GH26 domain from PbGH26-GH5A endo-beta-mannanase/endo-beta-glucanase from Prevotella bryantii [Prevotella bryantii B14] |
| 5D9N_A | 3.39e-144 | 423 | 780 | 2 | 352 | Crystalstructure of PbGH5A, a glycoside hydrolase family 5 member from Prevotella bryantii B14, in complex with the xyloglucan heptasaccharide XXXG [Prevotella bryantii],5D9N_B Crystal structure of PbGH5A, a glycoside hydrolase family 5 member from Prevotella bryantii B14, in complex with the xyloglucan heptasaccharide XXXG [Prevotella bryantii],5D9P_A Crystal structure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14, in complex with an inhibitory N-bromoacetylglycosylamine derivative of XXXG [Prevotella bryantii],5D9P_B Crystal structure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14, in complex with an inhibitory N-bromoacetylglycosylamine derivative of XXXG [Prevotella bryantii] |
| 5D9M_A | 2.69e-143 | 423 | 780 | 2 | 352 | Crystalstructure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14, E280A mutant in complex with the xyloglucan tetradecasaccharide XXXGXXXG [Prevotella bryantii],5D9M_B Crystal structure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14, E280A mutant in complex with the xyloglucan tetradecasaccharide XXXGXXXG [Prevotella bryantii],5D9O_A Crystal structure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14, E280A mutant in complex with cellotetraose [Prevotella bryantii],5D9O_B Crystal structure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14, E280A mutant in complex with cellotetraose [Prevotella bryantii] |
| 3VDH_A | 6.68e-138 | 423 | 780 | 2 | 352 | Crystalstructure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14 [Prevotella bryantii],3VDH_B Crystal structure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14 [Prevotella bryantii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P54937 | 1.99e-53 | 433 | 779 | 38 | 378 | Endoglucanase A OS=Clostridium longisporum OX=1523 GN=celA PE=1 SV=1 |
| Q12647 | 2.81e-53 | 433 | 752 | 24 | 328 | Endoglucanase B OS=Neocallimastix patriciarum OX=4758 GN=CELB PE=2 SV=1 |
| P17901 | 8.83e-51 | 438 | 777 | 51 | 398 | Endoglucanase A OS=Ruminiclostridium cellulolyticum (strain ATCC 35319 / DSM 5812 / JCM 6584 / H10) OX=394503 GN=celCCA PE=1 SV=1 |
| P28623 | 3.34e-49 | 434 | 777 | 43 | 370 | Endoglucanase D OS=Clostridium cellulovorans (strain ATCC 35296 / DSM 3052 / OCM 3 / 743B) OX=573061 GN=engD PE=1 SV=2 |
| P20847 | 4.39e-49 | 431 | 721 | 36 | 332 | Endoglucanase 1 OS=Butyrivibrio fibrisolvens OX=831 GN=end1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
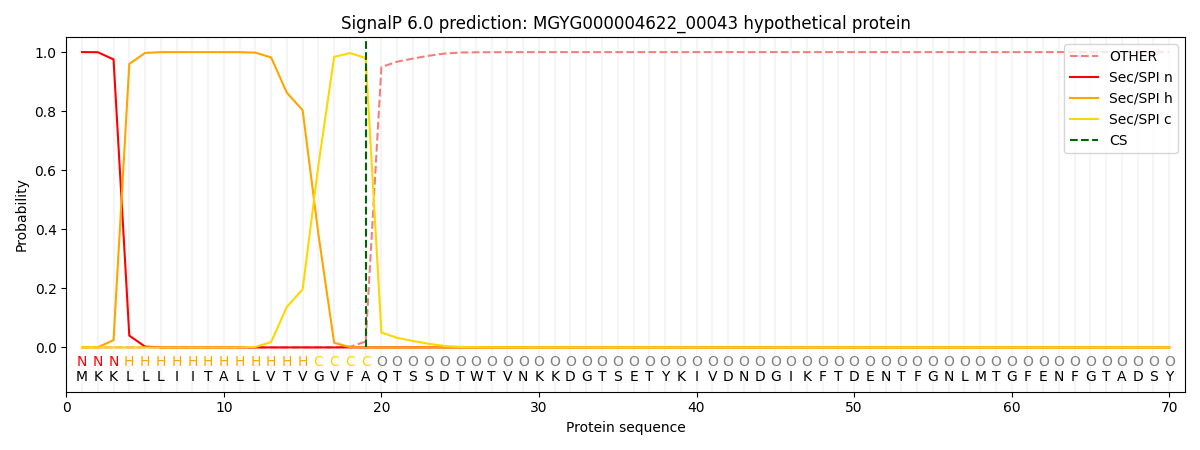
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000329 | 0.998912 | 0.000244 | 0.000164 | 0.000158 | 0.000154 |
