You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004621_01389
You are here: Home > Sequence: MGYG000004621_01389
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
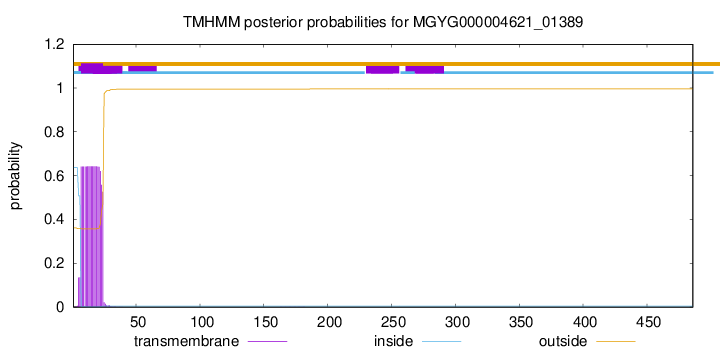
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; | |||||||||||
| CAZyme ID | MGYG000004621_01389 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 7661; End: 9121 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH18 | 27 | 327 | 1e-67 | 0.9594594594594594 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd06548 | GH18_chitinase | 4.95e-92 | 29 | 320 | 1 | 322 | The GH18 (glycosyl hydrolases, family 18) type II chitinases hydrolyze chitin, an abundant polymer of N-acetylglucosamine and have been identified in bacteria, fungi, insects, plants, viruses, and protozoan parasites. The structure of this domain is an eight-stranded alpha/beta barrel with a pronounced active-site cleft at the C-terminal end of the beta-barrel. |
| smart00636 | Glyco_18 | 3.16e-73 | 29 | 320 | 2 | 334 | Glyco_18 domain. |
| pfam00704 | Glyco_hydro_18 | 1.01e-71 | 29 | 320 | 2 | 307 | Glycosyl hydrolases family 18. |
| cd02872 | GH18_chitolectin_chitotriosidase | 5.41e-68 | 29 | 324 | 1 | 345 | This conserved domain family includes a large number of catalytically inactive chitinase-like lectins (chitolectins) including YKL-39, YKL-40 (HCGP39), YM1, oviductin, and AMCase (acidic mammalian chitinase), as well as catalytically active chitotriosidases. The conserved domain is an eight-stranded alpha/beta barrel fold belonging to the family 18 glycosyl hydrolases. The fold has a pronounced active-site cleft at the C-terminal end of the beta-barrel. The chitolectins lack a key active site glutamate (the proton donor required for hydrolytic activity) but retain highly conserved residues involved in oligosaccharide binding. Chitotriosidase is a chitinolytic enzyme expressed in maturing macrophages, which suggests that it plays a part in antimicrobial defense. Chitotriosidase hydrolyzes chitotriose, as well as colloidal chitin to yield chitobiose and is therefore considered an exochitinase. Chitotriosidase occurs in two major forms, the large form being converted to the small form by either RNA or post-translational processing. Although the small form, containing the chitinase domain alone, is sufficient for the chitinolytic activity, the additional C-terminal chitin-binding domain of the large form plays a role in processing colloidal chitin. The chitotriosidase gene is nonessential in humans, as about 35% of the population are heterozygous and 6% homozygous for an inactivated form of the gene. HCGP39 is a 39-kDa human cartilage glycoprotein thought to play a role in connective tissue remodeling and defense against pathogens. |
| COG3325 | ChiA | 3.01e-53 | 77 | 330 | 119 | 432 | Chitinase, GH18 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QYR11686.1 | 5.70e-208 | 21 | 481 | 20 | 508 |
| QUU00240.1 | 1.99e-139 | 21 | 440 | 29 | 456 |
| QUT62465.1 | 3.20e-139 | 21 | 440 | 22 | 449 |
| QQA31016.1 | 3.98e-139 | 21 | 440 | 29 | 456 |
| QBJ17604.1 | 5.63e-139 | 21 | 440 | 29 | 456 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6INX_A | 4.47e-49 | 24 | 316 | 2 | 325 | Structuralinsights into a novel glycoside hydrolase family 18 N-acetylglucosaminidase from Paenibacillus barengoltzii [Paenibacillus barengoltzii] |
| 6HM1_A | 8.85e-45 | 38 | 321 | 22 | 381 | Structuraland thermodynamic signatures of ligand binding to an enigmatic chitinase-D from Serratia proteamaculans [Serratia proteamaculans 568] |
| 4NZC_A | 1.64e-44 | 38 | 321 | 25 | 384 | Crystalstructure of Chitinase D from Serratia proteamaculans at 1.45 Angstrom resolution [Serratia proteamaculans 568],4Q22_A Crystal structure of Chitinase D from Serratia proteamaculans in complex with N-acetyl glucosamine at 1.93 Angstrom resolution [Serratia proteamaculans 568] |
| 4PTM_A | 1.82e-44 | 38 | 321 | 25 | 384 | CrystalStructure of Chitinase D from Serratia proteamaculans in complex with N-acetyl glucosamine, a hydrolyzed product of hexasaccharide at 1.7 Angstrom resolution [Serratia proteamaculans 568] |
| 4LGX_A | 1.94e-44 | 38 | 321 | 28 | 387 | Structureof Chitinase D from Serratia proteamaculans revealed an unusually constrained substrate binding site [Serratia proteamaculans 568] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P20533 | 6.56e-36 | 2 | 320 | 11 | 438 | Chitinase A1 OS=Niallia circulans OX=1397 GN=chiA1 PE=1 SV=1 |
| Q13231 | 3.53e-33 | 19 | 324 | 16 | 367 | Chitotriosidase-1 OS=Homo sapiens OX=9606 GN=CHIT1 PE=1 SV=1 |
| Q11174 | 3.15e-31 | 32 | 321 | 58 | 400 | Probable endochitinase OS=Caenorhabditis elegans OX=6239 GN=cht-1 PE=1 SV=1 |
| Q873X9 | 1.63e-30 | 72 | 335 | 115 | 404 | Endochitinase B1 OS=Neosartorya fumigata OX=746128 GN=chiB1 PE=1 SV=1 |
| E9QRF2 | 1.63e-30 | 72 | 335 | 115 | 404 | Endochitinase B1 OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=chiB1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
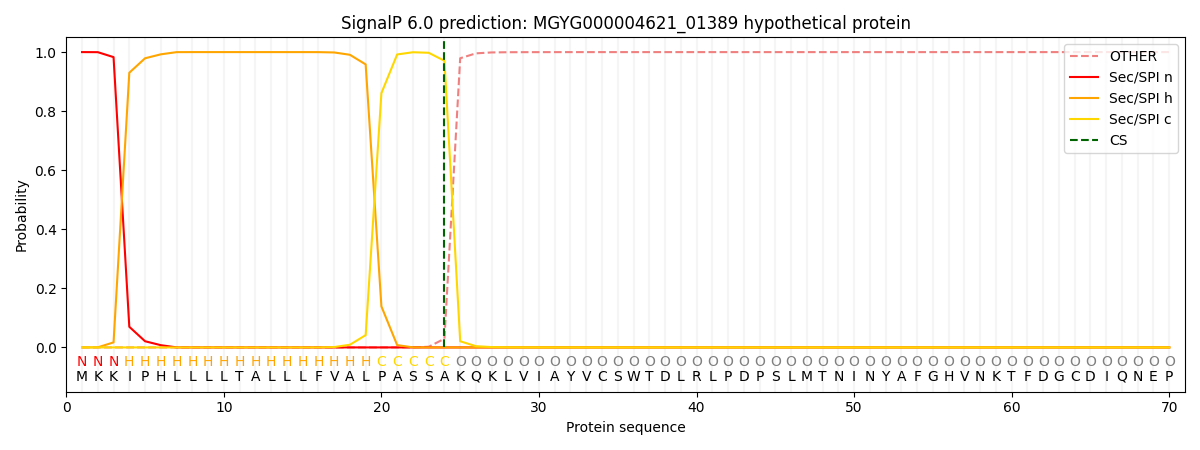
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000201 | 0.999227 | 0.000155 | 0.000143 | 0.000128 | 0.000130 |