You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004532_00957
You are here: Home > Sequence: MGYG000004532_00957
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Akkermansia sp900548895 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Verrucomicrobiales; Akkermansiaceae; Akkermansia; Akkermansia sp900548895 | |||||||||||
| CAZyme ID | MGYG000004532_00957 | |||||||||||
| CAZy Family | GH109 | |||||||||||
| CAZyme Description | Glycosyl hydrolase family 109 protein 1 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 14855; End: 16342 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH109 | 61 | 487 | 1.6e-159 | 0.9924812030075187 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0673 | MviM | 1.80e-20 | 65 | 422 | 4 | 330 | Predicted dehydrogenase [General function prediction only]. |
| pfam01408 | GFO_IDH_MocA | 7.15e-16 | 65 | 191 | 1 | 118 | Oxidoreductase family, NAD-binding Rossmann fold. This family of enzymes utilize NADP or NAD. This family is called the GFO/IDH/MOCA family in swiss-prot. |
| PRK11579 | PRK11579 | 1.57e-05 | 63 | 240 | 3 | 171 | putative oxidoreductase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| SEI00642.1 | 7.45e-245 | 32 | 494 | 35 | 474 |
| QHV64416.1 | 5.56e-220 | 46 | 495 | 56 | 481 |
| QHV71784.1 | 5.56e-220 | 46 | 495 | 56 | 481 |
| QHV69330.1 | 5.56e-220 | 46 | 495 | 56 | 481 |
| QHV66866.1 | 5.56e-220 | 46 | 495 | 56 | 481 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6T2B_A | 1.76e-199 | 39 | 494 | 17 | 447 | Glycosidehydrolase family 109 from Akkermansia muciniphila in complex with GalNAc and NAD+. [Akkermansia muciniphila],6T2B_B Glycoside hydrolase family 109 from Akkermansia muciniphila in complex with GalNAc and NAD+. [Akkermansia muciniphila],6T2B_C Glycoside hydrolase family 109 from Akkermansia muciniphila in complex with GalNAc and NAD+. [Akkermansia muciniphila],6T2B_D Glycoside hydrolase family 109 from Akkermansia muciniphila in complex with GalNAc and NAD+. [Akkermansia muciniphila] |
| 2IXA_A | 1.36e-76 | 65 | 486 | 21 | 432 | A-zyme,N-acetylgalactosaminidase [Elizabethkingia meningoseptica],2IXB_A Crystal structure of N-ACETYLGALACTOSAMINIDASE in complex with GalNAC [Elizabethkingia meningoseptica] |
| 3E18_A | 1.34e-17 | 61 | 221 | 2 | 151 | CRYSTALSTRUCTURE OF NAD-BINDING PROTEIN FROM Listeria innocua [Listeria innocua],3E18_B CRYSTAL STRUCTURE OF NAD-BINDING PROTEIN FROM Listeria innocua [Listeria innocua] |
| 3EC7_A | 3.80e-10 | 65 | 228 | 24 | 186 | CrystalStructure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_B Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_C Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_D Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_E Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_F Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_G Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_H Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2] |
| 1EVJ_A | 1.54e-08 | 68 | 216 | 6 | 150 | ChainA, GLUCOSE-FRUCTOSE OXIDOREDUCTASE [Zymomonas mobilis],1EVJ_B Chain B, GLUCOSE-FRUCTOSE OXIDOREDUCTASE [Zymomonas mobilis],1EVJ_C Chain C, GLUCOSE-FRUCTOSE OXIDOREDUCTASE [Zymomonas mobilis],1EVJ_D Chain D, GLUCOSE-FRUCTOSE OXIDOREDUCTASE [Zymomonas mobilis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B2UL75 | 6.40e-220 | 46 | 495 | 56 | 481 | Glycosyl hydrolase family 109 protein 1 OS=Akkermansia muciniphila (strain ATCC BAA-835 / DSM 22959 / JCM 33894 / BCRC 81048 / CCUG 64013 / CIP 107961 / Muc) OX=349741 GN=Amuc_0017 PE=3 SV=1 |
| B2UQL7 | 1.76e-198 | 39 | 494 | 42 | 472 | Glycosyl hydrolase family 109 protein 2 OS=Akkermansia muciniphila (strain ATCC BAA-835 / DSM 22959 / JCM 33894 / BCRC 81048 / CCUG 64013 / CIP 107961 / Muc) OX=349741 GN=Amuc_0920 PE=1 SV=1 |
| Q8ECL7 | 4.00e-169 | 40 | 494 | 30 | 457 | Alpha-N-acetylgalactosaminidase OS=Shewanella oneidensis (strain MR-1) OX=211586 GN=nagA PE=3 SV=1 |
| Q0HWR6 | 4.60e-168 | 40 | 494 | 30 | 457 | Glycosyl hydrolase family 109 protein 1 OS=Shewanella sp. (strain MR-7) OX=60481 GN=Shewmr7_1440 PE=3 SV=1 |
| Q0HKG4 | 4.60e-168 | 40 | 494 | 30 | 457 | Glycosyl hydrolase family 109 protein 1 OS=Shewanella sp. (strain MR-4) OX=60480 GN=Shewmr4_1375 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
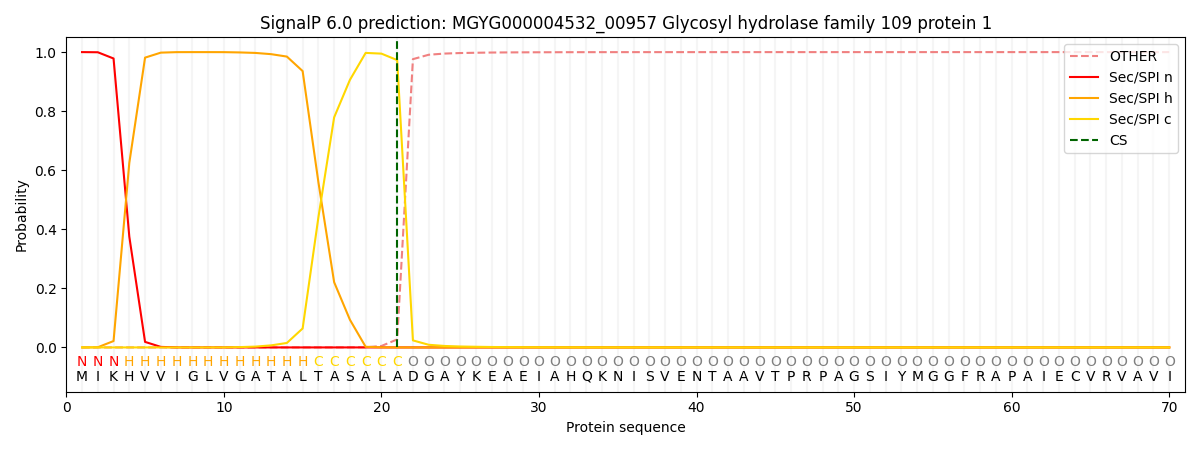
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000442 | 0.998752 | 0.000180 | 0.000227 | 0.000199 | 0.000171 |
