You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004490_01906
You are here: Home > Sequence: MGYG000004490_01906
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
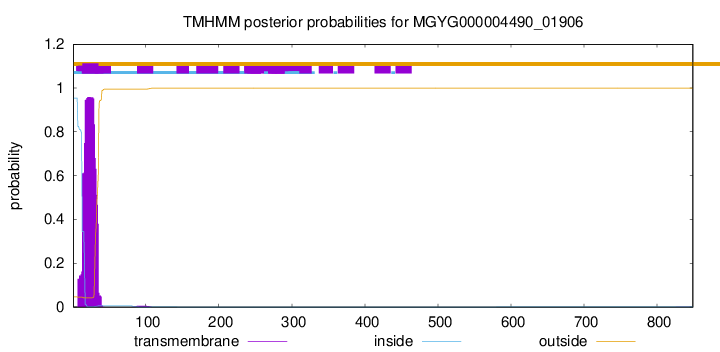
TMHMM annotations
Basic Information help
| Species | Muribaculum sp003150235 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; Muribaculum; Muribaculum sp003150235 | |||||||||||
| CAZyme ID | MGYG000004490_01906 | |||||||||||
| CAZy Family | GH115 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 29818; End: 32367 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH115 | 53 | 831 | 6.4e-212 | 0.9856527977044476 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam15979 | Glyco_hydro_115 | 0.0 | 181 | 518 | 1 | 334 | Glycosyl hydrolase family 115. Glyco_hydro_115 is a family of glycoside hydrolases likely to have the activity of xylan a-1,2-glucuronidase, EC:3.2.1.131, or a-(4-O-methyl)-glucuronidase EC:3.2.1.-. |
| pfam17829 | GH115_C | 1.68e-25 | 660 | 844 | 16 | 169 | Gylcosyl hydrolase family 115 C-terminal domain. This domain is found at the C-terminus of glycosyl hydrolase family 115 proteins. This domain has a beta-sandwich fold. |
| pfam03648 | Glyco_hydro_67N | 4.01e-04 | 83 | 148 | 57 | 120 | Glycosyl hydrolase family 67 N-terminus. Alpha-glucuronidases, components of an ensemble of enzymes central to the recycling of photosynthetic biomass, remove the alpha-1,2 linked 4-O-methyl glucuronic acid from xylans. This family represents the N-terminal region of alpha-glucuronidase. The N-terminal domain forms a two-layer sandwich, each layer being formed by a beta sheet of five strands. A further two helices form part of the interface with the central, catalytic, module (pfam07488). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRQ50360.1 | 1.59e-305 | 57 | 844 | 29 | 962 |
| EEC55419.1 | 3.67e-305 | 57 | 844 | 54 | 987 |
| QUT25621.1 | 3.56e-276 | 15 | 844 | 10 | 1003 |
| ALJ47135.1 | 1.78e-275 | 15 | 844 | 6 | 999 |
| SCV06546.1 | 2.02e-275 | 15 | 844 | 10 | 1003 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4C90_A | 4.56e-161 | 35 | 842 | 31 | 843 | Evidencethat GH115 alpha-glucuronidase activity is dependent on conformational flexibility [Bacteroides ovatus],4C90_B Evidence that GH115 alpha-glucuronidase activity is dependent on conformational flexibility [Bacteroides ovatus],4C91_A Evidence that GH115 alpha-glucuronidase activity is dependent on conformational flexibility [Bacteroides ovatus],4C91_B Evidence that GH115 alpha-glucuronidase activity is dependent on conformational flexibility [Bacteroides ovatus] |
| 4ZMH_A | 3.44e-159 | 34 | 641 | 1 | 637 | Crystalstructure of a five-domain GH115 alpha-Glucuronidase from the Marine Bacterium Saccharophagus degradans 2-40T [Saccharophagus degradans 2-40],4ZMH_B Crystal structure of a five-domain GH115 alpha-Glucuronidase from the Marine Bacterium Saccharophagus degradans 2-40T [Saccharophagus degradans 2-40] |
| 7PUG_A | 5.73e-154 | 58 | 679 | 28 | 695 | ChainA, xylan alpha-1,2-glucuronidase [uncultured bacterium] |
| 7PXQ_A | 1.84e-151 | 58 | 679 | 27 | 694 | ChainA, xylan alpha-1,2-glucuronidase [uncultured bacterium] |
| 6NPS_A | 1.10e-115 | 41 | 634 | 5 | 654 | Crystalstructure of GH115 enzyme AxyAgu115A from Amphibacillus xylanus [Amphibacillus xylanus NBRC 15112],6NPS_B Crystal structure of GH115 enzyme AxyAgu115A from Amphibacillus xylanus [Amphibacillus xylanus NBRC 15112] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
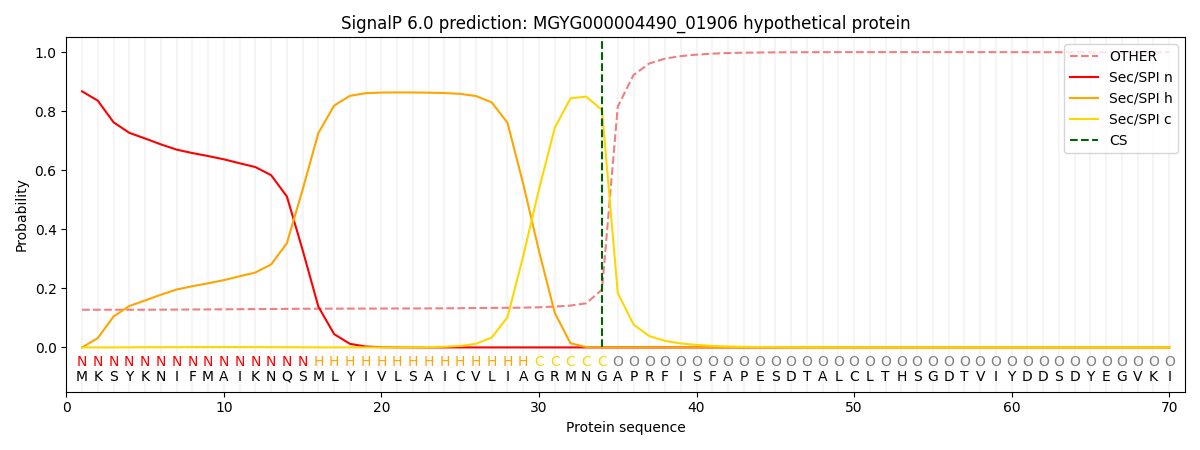
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.133179 | 0.859090 | 0.006637 | 0.000447 | 0.000282 | 0.000333 |