You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004429_00834
You are here: Home > Sequence: MGYG000004429_00834
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
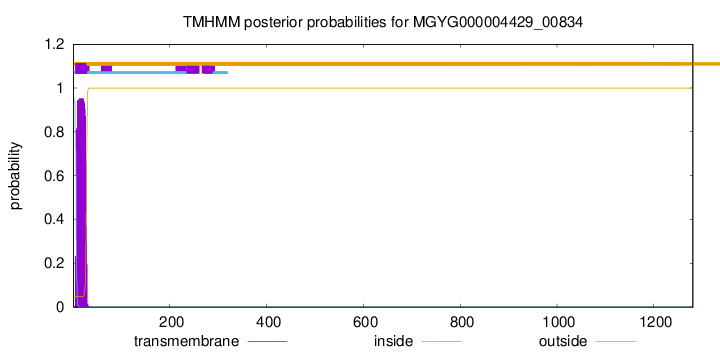
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; CAG-274; ; | |||||||||||
| CAZyme ID | MGYG000004429_00834 | |||||||||||
| CAZy Family | GH98 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1266; End: 5111 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH98 | 61 | 386 | 2.6e-135 | 0.9877675840978594 |
| CE1 | 1047 | 1276 | 3e-41 | 0.973568281938326 |
| CBM35 | 678 | 771 | 1.1e-20 | 0.7899159663865546 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08306 | Glyco_hydro_98M | 7.34e-152 | 63 | 386 | 8 | 328 | Glycosyl hydrolase family 98. This domain is the putative catalytic domain of glycosyl hydrolase family 98 proteins. |
| cd04083 | CBM35_Lmo2446-like | 1.57e-31 | 656 | 784 | 1 | 125 | Carbohydrate Binding Module 35 (CBM35) domains similar to Lmo2446. This family includes carbohydrate binding module 35 (CBM35) domains that are appended to several carbohydrate binding enzymes. Some CBM35 domains belonging to this family are appended to glycoside hydrolase (GH) family domains, including glycoside hydrolase family 31 (GH31), for example the CBM35 domain of Lmo2446, an uncharacterized protein from Listeria monocytogenes EGD-e. These CBM35s are non-catalytic carbohydrate binding domains that facilitate the strong binding of the GH catalytic modules with their dedicated, insoluble substrates. GH31 has a wide range of hydrolytic activities such as alpha-glucosidase, alpha-xylosidase, 6-alpha-glucosyltransferase, or alpha-1,4-glucan lyase, cleaving a terminal carbohydrate moiety from a substrate that may be a starch or a glycoprotein. Most characterized GH31 enzymes are alpha-glucosidases. |
| COG2382 | Fes | 8.02e-27 | 1013 | 1281 | 42 | 299 | Enterochelin esterase or related enzyme [Inorganic ion transport and metabolism]. |
| COG2819 | YbbA | 1.11e-17 | 1057 | 1199 | 26 | 171 | Predicted hydrolase of the alpha/beta superfamily [General function prediction only]. |
| pfam00756 | Esterase | 2.52e-16 | 1051 | 1198 | 5 | 143 | Putative esterase. This family contains Esterase D. However it is not clear if all members of the family have the same function. This family is related to the pfam00135 family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBL18082.1 | 0.0 | 4 | 1027 | 4 | 1031 |
| ADL51037.1 | 0.0 | 4 | 810 | 4 | 806 |
| BAV13045.1 | 0.0 | 4 | 810 | 4 | 806 |
| ADU21039.1 | 0.0 | 50 | 942 | 32 | 935 |
| AOZ99817.1 | 8.74e-240 | 48 | 781 | 22 | 738 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5CXU_A | 1.50e-56 | 1025 | 1280 | 11 | 272 | Structureof the CE1 ferulic acid esterase AmCE1/Fae1A, from the anaerobic fungi Anaeromyces mucronatus in the absence of substrate [Anaeromyces mucronatus],5CXX_A Structure of a CE1 ferulic acid esterase, AmCE1/Fae1A, from Anaeromyces mucronatus in complex with Ferulic acid [Anaeromyces mucronatus],5CXX_B Structure of a CE1 ferulic acid esterase, AmCE1/Fae1A, from Anaeromyces mucronatus in complex with Ferulic acid [Anaeromyces mucronatus],5CXX_C Structure of a CE1 ferulic acid esterase, AmCE1/Fae1A, from Anaeromyces mucronatus in complex with Ferulic acid [Anaeromyces mucronatus] |
| 1JJF_A | 9.74e-33 | 1017 | 1280 | 9 | 260 | ChainA, Endo-1,4-beta-xylanase Z [Acetivibrio thermocellus] |
| 1JT2_A | 2.44e-32 | 1017 | 1280 | 9 | 260 | ChainA, PROTEIN (ENDO-1,4-BETA-XYLANASE Z) [Acetivibrio thermocellus] |
| 6RZN_A | 1.86e-23 | 1020 | 1279 | 112 | 386 | Crystalstructure of the N-terminal carbohydrate binding module family 48 and ferulic acid esterase from the multi-enzyme CE1-GH62-GH10 [uncultured bacterium],6RZN_B Crystal structure of the N-terminal carbohydrate binding module family 48 and ferulic acid esterase from the multi-enzyme CE1-GH62-GH10 [uncultured bacterium] |
| 2WMF_A | 6.77e-19 | 172 | 625 | 134 | 581 | Crystalstructure of the catalytic module of a family 98 glycoside hydrolase from Streptococcus pneumoniae TIGR4 (Sp4GH98) in its native form. [Streptococcus pneumoniae TIGR4] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P10478 | 3.79e-29 | 1017 | 1280 | 28 | 279 | Endo-1,4-beta-xylanase Z OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=xynZ PE=1 SV=3 |
| D5EXZ4 | 2.44e-14 | 1020 | 1232 | 411 | 626 | Carbohydrate acetyl esterase/feruloyl esterase OS=Prevotella ruminicola (strain ATCC 19189 / JCM 8958 / 23) OX=264731 GN=axe1-6A PE=1 SV=1 |
| P51584 | 8.86e-13 | 948 | 1235 | 733 | 1022 | Endo-1,4-beta-xylanase Y OS=Acetivibrio thermocellus OX=1515 GN=xynY PE=1 SV=1 |
| Q56F26 | 7.79e-06 | 634 | 771 | 885 | 1020 | Exo-beta-D-glucosaminidase OS=Amycolatopsis orientalis OX=31958 GN=csxA PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000347 | 0.998991 | 0.000186 | 0.000174 | 0.000151 | 0.000137 |