You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004424_00047
You are here: Home > Sequence: MGYG000004424_00047
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
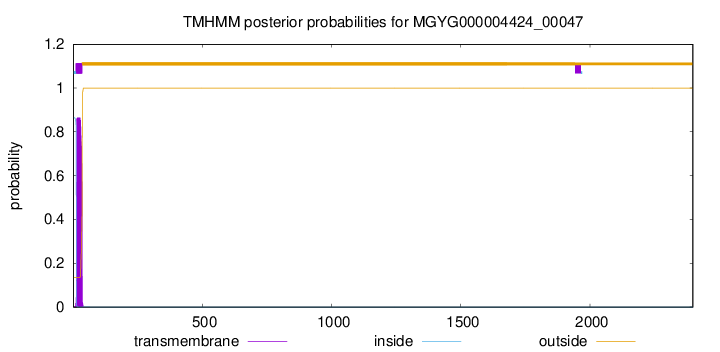
TMHMM annotations
Basic Information help
| Species | Collinsella sp900548365 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Coriobacteriia; Coriobacteriales; Coriobacteriaceae; Collinsella; Collinsella sp900548365 | |||||||||||
| CAZyme ID | MGYG000004424_00047 | |||||||||||
| CAZy Family | GH31 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 70167; End: 77363 Strand: - | |||||||||||
Full Sequence Download help
| MGYGIPKHWK MGGVGFVLAT SLAAGWALNP ALATPAFADE AMASDYQGTL TSATAAKKDA | 60 |
| DNDNIAYVTF NDNVTAKITF LEPGIFRYNV DLSGDFSAYA TPRAKSHTAK IQAQPDTSGK | 120 |
| YSKPAATVNE TKDAFEVSDG TVTLSFEKAT GKMTLKRGDR VVFAEDEPLT ITKSATTQSI | 180 |
| ADADADYFGG GTQNGRFIHT GKAINIKNES NWVDGGVSSP NPFYWSTAGY GVMRNTFAEG | 240 |
| KYDFGSAVAG SVDALHKDGE FDAYYFVSDD ADTTASVAQD VLQEYYKVTG SPLLLPEYAF | 300 |
| YVGNYNAYNR DAWSHEPKSG YKEWSIHGHE SASKAAPSKR YEKGGTGQTM LANSYVESLN | 360 |
| GTAPKDAETN ENIPEGVQWS EDFSARAVVD EYQDMDMPFG YILPNDGYGC GYGQNGYQKT | 420 |
| GGVDKDGNSS AERLQAVADN VQNLKEFSDY AKSKGVATGL WTQSNLSPDS NANTQWQTLR | 480 |
| DFESEVKKGG VTTLKTDVAW VGSGYSFQLN GTKTAYDIVT TKNGDGTGAR PNIVSLDGWA | 540 |
| GSQRFAGLWS GDQTGGNWEY IRFHIPTFIG SGLSGNPNIG SDMDGIFGGH PLIACRDYQW | 600 |
| KSFSSLMLDM DGWGSYAKMP YAYGDPYTGI NRMYLKLKSS LLPYIYTTAA SAANIDTGNG | 660 |
| DEGLPIVRAI ALSDNSDIAN STATQYEYTL GEDLLIAPVY QNTDGDSANG GLGDGDDIRN | 720 |
| DIYLPGTSED IWIDYWTGDQ YRGGQVLNNF AVPLWKTPVF VKANAIIPMY KPNDNPSDID | 780 |
| RAQRDIEFFA TDGENEYTLY EDDGSYVENK IDESDKEYGR ESTISYGDHV STKITSAVKD | 840 |
| GTATFTAAKS TGGYDGYDAN RTTTFVVNVS AKPSELVAKN GDKTLELNEV ASKEDFDKAE | 900 |
| GNVYFYNEAP NLNYNATAED EAVRNEEFSK TEITTTPKLY VKFAKTDVTK DAQTLTLKGF | 960 |
| ENKGDLPANS LNENLAAPAN LAAPEDDITP TSIKLTWDAV AGATGYELEL DGVLSSVGDT | 1020 |
| TTFTHANLAY NSSHTYRVRA VNADGYSAWS EPLTTKTALD PWRNVPVPVD WDFEGSEFGS | 1080 |
| GYGVKFAFDH ITGADASSFV SKETNGTGLA LDLDYGKVYQ FEKLEFRGSK YGQGVKQMKI | 1140 |
| EASLDGTHWT DLGTHDLSGS FDKLNTVEFG APVSARYIRM TAVQCTSYWN ATEIAFYKVD | 1200 |
| GINGAELGSI NGDAVVDSAD YQHLTGNCLG RENREPEAAS YQTHVAKNGA DFNQNGAYDV | 1260 |
| YDMAFTMSKL DGGTTKTGDV SGGIAVMPSA THVEAGDTIT VDVYASDAKN VNALGALVHF | 1320 |
| KSDQFEFVKD SIEQSPYTST MENLSIAKTE FDDGIQSVNL AFANKGDKEL YDGSGVVASF | 1380 |
| KLKAVAAGDV NLDSTAWLIG PTNDSIDVVS DGTIDWPEIP GEHEEEYAQS AFNCSILNEK | 1440 |
| GEAVDVSKYI HQENFDGLFN GDVNSNDFEM EWQNAGDFDT SFHKLPATLR FEFKKPSALE | 1500 |
| NVVVYNRTSG NGCVTELDAS IVFEDGTKQD FTFGAKQNTF ELAVSQENAG KKVARVDITP | 1560 |
| KNTTTGINML TLREIDFTYT TPGAQVAGVT VDDQTQTELY QGDLAQVFAT VDCDDYPYFE | 1620 |
| VSSDKPEVAS VTAVQSGEGV DWYVRGNAEG TATITVAAKA DPSKTATYEV TVRAGVDVSG | 1680 |
| LQAIISEGRI YDSEAYTEAS FAKLEAALKA AEDMLAQGQG SFTKNDVAQK SMDIENAIKG | 1740 |
| LKMRPIDEAK LLNKDASSGM SVASVSSYAG ESPMELALDY DEDTLWHSNY GSSMRLPQYI | 1800 |
| VYDLGAEYDL TDVTFLPRQN GSLNGDIFKA QVYVADSVDE LTAAADSGTL VGTFSFDNNG | 1860 |
| KTLTNRNEYQ QMAFGATTTR YVKINVIESG SSDGAGNRYC SIAETRFYGE KHTDPIGDAK | 1920 |
| AELSTKVAEY EAKGLNADDY TASTWLPYAN ALADAKSAIE GDGLTAEQIA AVGERLDTAF | 1980 |
| AGLKKTEIPP VVDADKEKLQ AVVDLVKDTD LDGKTEATAK RFGDALKAAQ QALKAGEGNW | 2040 |
| KALYDELKAA YDGLADEAEV DTATLQGFVN MFDSLGMTSA DFTADSWKAY ADALAAAKDV | 2100 |
| LAKDDATQEQ VNACTDALTS AFKGLEFKQA PAAPYKGYLQ KVVANFASEG LDESRYTADS | 2160 |
| WKAYADALKA AQGVLDADDA TQEQIDDATT ALVEAHAGLQ PAEKISFSDV DATVSHQDDI | 2220 |
| VWLAANGISK GWENADGTFS FHPYENVARA DMAAFLYRMA GEPEVDAEKA PSFTDVEKDT | 2280 |
| PHYKAILWLA AEGISTGWEN ADGTAEFRPY AQITRADMAA FLYRMAGKPD VESPELGGFT | 2340 |
| DVDEGTVHSD AILWLAAEGI STGWEHEDGT AEFRPYDQIT RADMAAFLHR MDQKGLVK | 2398 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH31 | 511 | 767 | 1.3e-47 | 0.531615925058548 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd06596 | GH31_CPE1046 | 5.66e-134 | 288 | 738 | 1 | 334 | Clostridium CPE1046-like. CPE1046 is an uncharacterized Clostridium perfringens protein with a glycosyl hydrolase family 31 (GH31) domain. The domain architecture of CPE1046 and its orthologs includes a C-terminal fibronectin type 3 (FN3) domain and a coagulation factor 5/8 type C domain in addition to the GH31 domain. Enzymes of the GH31 family possess a wide range of different hydrolytic activities including alpha-glucosidase (glucoamylase and sucrase-isomaltase), alpha-xylosidase, 6-alpha-glucosyltransferase, 3-alpha-isomaltosyltransferase and alpha-1,4-glucan lyase. All GH31 enzymes cleave a terminal carbohydrate moiety from a substrate that varies considerably in size, depending on the enzyme, and may be either a starch or a glycoprotein. |
| COG1501 | YicI | 6.12e-51 | 222 | 815 | 195 | 719 | Alpha-glucosidase, glycosyl hydrolase family GH31 [Carbohydrate transport and metabolism]. |
| pfam01055 | Glyco_hydro_31 | 1.26e-43 | 445 | 767 | 159 | 442 | Glycosyl hydrolases family 31. Glycosyl hydrolases are key enzymes of carbohydrate metabolism. Family 31 comprises of enzymes that are, or similar to, alpha- galactosidases. |
| cd08759 | Type_III_cohesin_like | 2.18e-34 | 1282 | 1404 | 1 | 131 | Cohesin domain, interaction partner of dockerin. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. Two specific calcium-dependent interactions between cohesin and dockerin appear to be essential for cellulosome assembly, type I and type II. This subfamily represents type III cohesins and closely related domains. |
| cd06589 | GH31 | 2.84e-30 | 384 | 635 | 25 | 265 | glycosyl hydrolase family 31 (GH31). GH31 enzymes occur in prokaryotes, eukaryotes, and archaea with a wide range of hydrolytic activities, including alpha-glucosidase (glucoamylase and sucrase-isomaltase), alpha-xylosidase, 6-alpha-glucosyltransferase, 3-alpha-isomaltosyltransferase and alpha-1,4-glucan lyase. All GH31 enzymes cleave a terminal carbohydrate moiety from a substrate that varies considerably in size, depending on the enzyme, and may be either a starch or a glycoprotein. In most cases, the pyranose moiety recognized in subsite -1 of the substrate binding site is an alpha-D-glucose, though some GH31 family members show a preference for alpha-D-xylose. Several GH31 enzymes can accommodate both glucose and xylose and different levels of discrimination between the two have been observed. Most characterized GH31 enzymes are alpha-glucosidases. In mammals, GH31 members with alpha-glucosidase activity are implicated in at least three distinct biological processes. The lysosomal acid alpha-glucosidase (GAA) is essential for glycogen degradation and a deficiency or malfunction of this enzyme causes glycogen storage disease II, also known as Pompe disease. In the endoplasmic reticulum, alpha-glucosidase II catalyzes the second step in the N-linked oligosaccharide processing pathway that constitutes part of the quality control system for glycoprotein folding and maturation. The intestinal enzymes sucrase-isomaltase (SI) and maltase-glucoamylase (MGAM) play key roles in the final stage of carbohydrate digestion, making alpha-glucosidase inhibitors useful in the treatment of type 2 diabetes. GH31 alpha-glycosidases are retaining enzymes that cleave their substrates via an acid/base-catalyzed, double-displacement mechanism involving a covalent glycosyl-enzyme intermediate. Two aspartic acid residues have been identified as the catalytic nucleophile and the acid/base, respectively. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QWT17625.1 | 0.0 | 46 | 2083 | 46 | 2128 |
| BCT46261.1 | 0.0 | 43 | 2131 | 36 | 2121 |
| QNM10857.1 | 0.0 | 43 | 2029 | 37 | 1983 |
| BBK61154.1 | 0.0 | 50 | 2118 | 39 | 2129 |
| BBK23937.1 | 0.0 | 64 | 2071 | 69 | 2012 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6M76_A | 2.65e-222 | 68 | 1063 | 52 | 963 | GH31alpha-N-acetylgalactosaminidase from Enterococcus faecalis [Enterococcus faecalis ATCC 10100],6M77_A GH31 alpha-N-acetylgalactosaminidase from Enterococcus faecalis in complex with N-acetylgalactosamine [Enterococcus faecalis ATCC 10100] |
| 7F7R_A | 1.37e-221 | 68 | 1063 | 52 | 963 | ChainA, GH31 alpha-N-acetylgalactosaminidase [Enterococcus faecalis ATCC 10100] |
| 7F7Q_A | 3.66e-221 | 68 | 1063 | 52 | 963 | ChainA, GH31 alpha-N-acetylgalactosaminidase [Enterococcus faecalis ATCC 10100] |
| 5F7C_A | 1.19e-17 | 513 | 778 | 488 | 726 | Crystalstructure of Family 31 alpha-glucosidase (BT_0339) from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482],5F7C_B Crystal structure of Family 31 alpha-glucosidase (BT_0339) from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482],5F7C_C Crystal structure of Family 31 alpha-glucosidase (BT_0339) from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482],5F7C_D Crystal structure of Family 31 alpha-glucosidase (BT_0339) from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482] |
| 7KBJ_A | 2.17e-15 | 483 | 770 | 198 | 466 | ChainA, Neutral alpha-glucosidase AB Trypsin-cleaved Fragment #3 [Mus musculus],7KBJ_C Chain C, Neutral alpha-glucosidase AB Trypsin-cleaved Fragment #3 [Mus musculus],7KBR_A Chain A, Neutral alpha-glucosidase AB Trypsin-cleaved Fragment #3 [Mus musculus],7KBR_C Chain C, Neutral alpha-glucosidase AB Trypsin-cleaved Fragment #3 [Mus musculus],7L9E_A Chain A, Neutral alpha-glucosidase AB Trypsin-cleaved Fragment #3 [Mus musculus],7L9E_C Chain C, Neutral alpha-glucosidase AB Trypsin-cleaved Fragment #3 [Mus musculus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9P999 | 3.22e-21 | 501 | 804 | 376 | 658 | Alpha-xylosidase OS=Saccharolobus solfataricus (strain ATCC 35092 / DSM 1617 / JCM 11322 / P2) OX=273057 GN=xylS PE=1 SV=1 |
| Q9F234 | 1.27e-19 | 528 | 804 | 460 | 711 | Alpha-glucosidase 2 OS=Bacillus thermoamyloliquefaciens OX=1425 PE=3 SV=1 |
| B9F676 | 1.99e-17 | 483 | 768 | 513 | 778 | Probable glucan 1,3-alpha-glucosidase OS=Oryza sativa subsp. japonica OX=39947 GN=Os03g0216600 PE=3 SV=1 |
| P79403 | 7.91e-17 | 483 | 770 | 545 | 813 | Neutral alpha-glucosidase AB OS=Sus scrofa OX=9823 GN=GANAB PE=1 SV=1 |
| Q4R4N7 | 9.09e-16 | 483 | 770 | 545 | 813 | Neutral alpha-glucosidase AB OS=Macaca fascicularis OX=9541 GN=GANAB PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
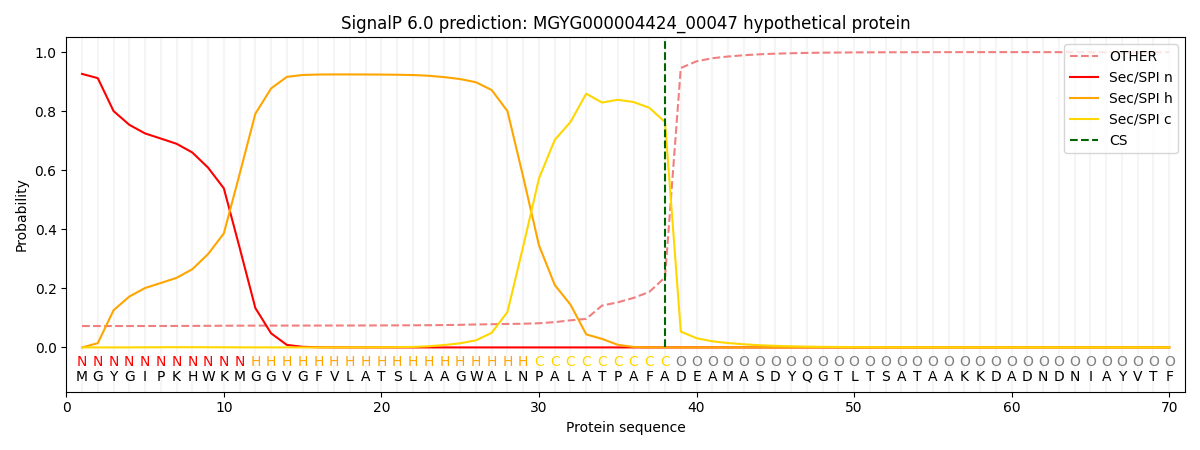
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.077516 | 0.919526 | 0.001749 | 0.000511 | 0.000343 | 0.000331 |