You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004132_02028
You are here: Home > Sequence: MGYG000004132_02028
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | V9D3004 sp900760345 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; V9D3004; V9D3004 sp900760345 | |||||||||||
| CAZyme ID | MGYG000004132_02028 | |||||||||||
| CAZy Family | CE1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 12264; End: 13664 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE1 | 207 | 460 | 1.4e-32 | 0.9515418502202643 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00756 | Esterase | 4.54e-13 | 213 | 451 | 11 | 235 | Putative esterase. This family contains Esterase D. However it is not clear if all members of the family have the same function. This family is related to the pfam00135 family. |
| COG2382 | Fes | 2.45e-12 | 166 | 361 | 49 | 210 | Enterochelin esterase or related enzyme [Inorganic ion transport and metabolism]. |
| NF033201 | Vip_LPXTG_Lm | 6.39e-05 | 105 | 205 | 256 | 352 | cell invasion LPXTG protein Vip. Vip (Virulence protein), like the LPXTG-type internalins, is an LPXTG-anchored surface protein of the mammalian cell-invading pathogen Listeria monocytogenes, but absent from the related species Listeria innocua. For certain cell types, Vip is required for Listeria's ability to invade. It appears to bind the endoplasmic reticulum (ER) resident chaperone Gp96 as its receptor. |
| NF033761 | gliding_GltJ | 7.16e-05 | 117 | 192 | 416 | 483 | adventurous gliding motility protein GltJ. Adventurous gliding motility protein GltJ, also known as AgmX, occurs in delta-proteobacteria such as Myxococcus xanthus. |
| NF033761 | gliding_GltJ | 1.73e-04 | 115 | 192 | 431 | 506 | adventurous gliding motility protein GltJ. Adventurous gliding motility protein GltJ, also known as AgmX, occurs in delta-proteobacteria such as Myxococcus xanthus. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AEY68182.1 | 6.33e-38 | 171 | 404 | 407 | 630 |
| AGA59284.1 | 9.80e-37 | 161 | 466 | 306 | 598 |
| AQR94781.1 | 1.06e-36 | 171 | 451 | 324 | 593 |
| ADL33750.1 | 2.81e-36 | 167 | 463 | 736 | 995 |
| AKA87411.1 | 5.42e-36 | 202 | 463 | 11 | 239 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1GKL_A | 4.10e-34 | 174 | 404 | 23 | 233 | ChainA, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],1GKL_B Chain B, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],1WB4_A Chain A, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus],1WB4_B Chain B, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus],1WB5_A Chain A, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus],1WB5_B Chain B, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus],1WB6_A Chain A, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],1WB6_B Chain B, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus] |
| 6FJ4_A | 1.03e-31 | 174 | 404 | 9 | 219 | ChainA, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus] |
| 1GKK_A | 1.40e-31 | 174 | 404 | 23 | 233 | ChainA, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus],1GKK_B Chain B, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus],3ZI7_A Chain A, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus YS],3ZI7_B Chain B, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus YS],4BAG_A Chain A, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],4BAG_B Chain B, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],4H35_A Chain A, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus],4H35_B Chain B, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus],5FXM_A Chain A, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus],6Y8G_AAA Chain AAA, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus],6Y8G_BBB Chain BBB, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus] |
| 1JJF_A | 4.74e-25 | 167 | 429 | 12 | 243 | ChainA, Endo-1,4-beta-xylanase Z [Acetivibrio thermocellus] |
| 1JT2_A | 1.22e-24 | 167 | 429 | 12 | 243 | ChainA, PROTEIN (ENDO-1,4-BETA-XYLANASE Z) [Acetivibrio thermocellus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P51584 | 3.72e-31 | 174 | 404 | 811 | 1021 | Endo-1,4-beta-xylanase Y OS=Acetivibrio thermocellus OX=1515 GN=xynY PE=1 SV=1 |
| P10478 | 1.05e-22 | 167 | 429 | 31 | 262 | Endo-1,4-beta-xylanase Z OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=xynZ PE=1 SV=3 |
| D5EY13 | 3.47e-13 | 170 | 434 | 480 | 715 | Endo-1,4-beta-xylanase/feruloyl esterase OS=Prevotella ruminicola (strain ATCC 19189 / JCM 8958 / 23) OX=264731 GN=xyn10D-fae1A PE=1 SV=1 |
| P31471 | 1.16e-07 | 171 | 361 | 128 | 304 | Uncharacterized protein YieL OS=Escherichia coli (strain K12) OX=83333 GN=yieL PE=4 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as SP
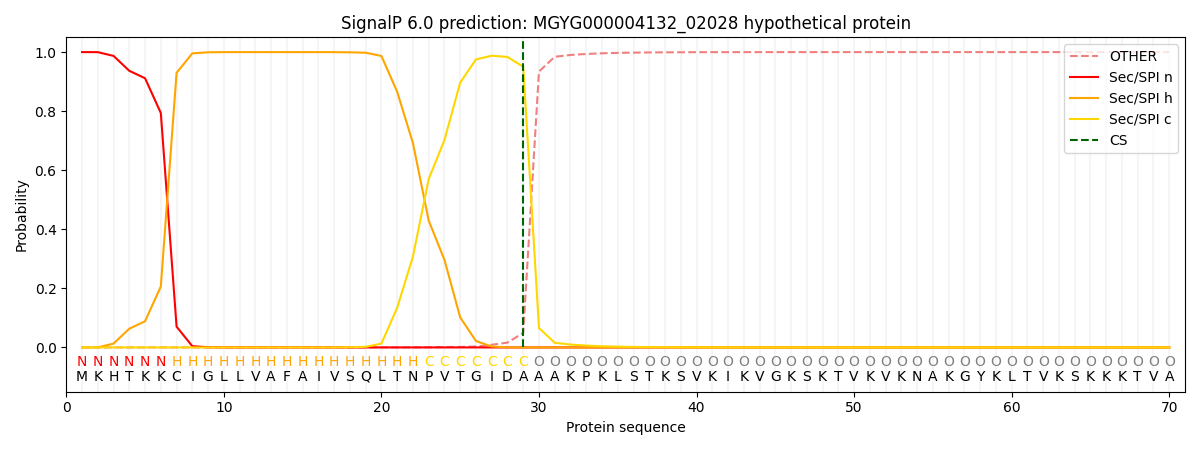
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000356 | 0.998953 | 0.000190 | 0.000167 | 0.000163 | 0.000148 |
