You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004127_00294
You are here: Home > Sequence: MGYG000004127_00294
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Propionibacteriales; Propionibacteriaceae; ; | |||||||||||
| CAZyme ID | MGYG000004127_00294 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Beta-hexosaminidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1225; End: 2415 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 125 | 348 | 1.9e-39 | 0.9583333333333334 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1472 | BglX | 2.60e-60 | 67 | 394 | 3 | 319 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| pfam00933 | Glyco_hydro_3 | 2.14e-44 | 67 | 386 | 2 | 316 | Glycosyl hydrolase family 3 N terminal domain. |
| PRK05337 | PRK05337 | 1.08e-35 | 75 | 386 | 4 | 305 | beta-hexosaminidase; Provisional |
| PRK15098 | PRK15098 | 9.16e-06 | 179 | 319 | 150 | 287 | beta-glucosidase BglX. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIK71752.1 | 1.43e-114 | 60 | 395 | 68 | 403 |
| QTE25615.1 | 4.24e-100 | 62 | 395 | 70 | 403 |
| BAK37793.1 | 7.53e-100 | 60 | 395 | 98 | 438 |
| SDE70461.1 | 1.24e-98 | 62 | 395 | 8 | 348 |
| QTO37029.1 | 3.96e-97 | 60 | 395 | 66 | 404 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5BU9_A | 8.04e-72 | 65 | 387 | 5 | 337 | Crystalstructure of Beta-N-acetylhexosaminidase from Beutenbergia cavernae DSM 12333 [Beutenbergia cavernae DSM 12333],5BU9_B Crystal structure of Beta-N-acetylhexosaminidase from Beutenbergia cavernae DSM 12333 [Beutenbergia cavernae DSM 12333] |
| 6K5J_A | 5.65e-31 | 63 | 388 | 9 | 338 | Structureof a glycoside hydrolase family 3 beta-N-acetylglucosaminidase from Paenibacillus sp. str. FPU-7 [Paenibacillaceae] |
| 3TEV_A | 5.23e-26 | 60 | 346 | 8 | 289 | Thecrystal structure of glycosyl hydrolase from Deinococcus radiodurans R1 [Deinococcus radiodurans R1],3TEV_B The crystal structure of glycosyl hydrolase from Deinococcus radiodurans R1 [Deinococcus radiodurans R1] |
| 4GVF_A | 1.31e-25 | 75 | 348 | 4 | 277 | Crystalstructure of Salmonella typhimurium family 3 glycoside hydrolase (NagZ) bound to GlcNAc [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],4GVF_B Crystal structure of Salmonella typhimurium family 3 glycoside hydrolase (NagZ) bound to GlcNAc [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],4GVG_A Crystal structure of Salmonella typhimurium family 3 glycoside hydrolase (NagZ) [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],4GVG_B Crystal structure of Salmonella typhimurium family 3 glycoside hydrolase (NagZ) [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],4GVH_A Crystal structure of Salmonella typhimurium family 3 glycoside hydrolase (NagZ) covalently bound to 5-fluoro-GlcNAc. [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],4GVH_B Crystal structure of Salmonella typhimurium family 3 glycoside hydrolase (NagZ) covalently bound to 5-fluoro-GlcNAc. [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],4HZM_A Crystal structure of Salmonella typhimurium family 3 glycoside hydrolase (NagZ) bound to N-[(3S,4R,5R,6R)-4,5-dihydroxy-6-(hydroxymethyl)piperidin-3-yl]butanamide [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],4HZM_B Crystal structure of Salmonella typhimurium family 3 glycoside hydrolase (NagZ) bound to N-[(3S,4R,5R,6R)-4,5-dihydroxy-6-(hydroxymethyl)piperidin-3-yl]butanamide [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2] |
| 5G1M_A | 4.87e-25 | 75 | 370 | 25 | 315 | Crystalstructure of NagZ from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],5G1M_B Crystal structure of NagZ from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],5G2M_A Crystal structure of NagZ from Pseudomonas aeruginosa in complex with N-acetylglucosamine [Pseudomonas aeruginosa PAO1],5G2M_B Crystal structure of NagZ from Pseudomonas aeruginosa in complex with N-acetylglucosamine [Pseudomonas aeruginosa PAO1],5G3R_A Crystal Structure Of Nagz From Pseudomonas Aeruginosa In Complex With N-acetylglucosamine And L-ala-1,6-anhydromurnac [Pseudomonas aeruginosa PAO1],5G3R_B Crystal Structure Of Nagz From Pseudomonas Aeruginosa In Complex With N-acetylglucosamine And L-ala-1,6-anhydromurnac [Pseudomonas aeruginosa PAO1],5G5K_A Crystal structure of NagZ from Pseudomonas aeruginosa in complex with the inhibitor 2-acetamido-1,2-dideoxynojirimycin [Pseudomonas aeruginosa PAO1],5G5K_B Crystal structure of NagZ from Pseudomonas aeruginosa in complex with the inhibitor 2-acetamido-1,2-dideoxynojirimycin [Pseudomonas aeruginosa PAO1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B5RB93 | 6.36e-25 | 75 | 348 | 4 | 277 | Beta-hexosaminidase OS=Salmonella gallinarum (strain 287/91 / NCTC 13346) OX=550538 GN=nagZ PE=3 SV=1 |
| B4TTH9 | 6.36e-25 | 75 | 348 | 4 | 277 | Beta-hexosaminidase OS=Salmonella schwarzengrund (strain CVM19633) OX=439843 GN=nagZ PE=3 SV=1 |
| B5F8E5 | 6.36e-25 | 75 | 348 | 4 | 277 | Beta-hexosaminidase OS=Salmonella agona (strain SL483) OX=454166 GN=nagZ PE=3 SV=1 |
| B5QXC7 | 6.36e-25 | 75 | 348 | 4 | 277 | Beta-hexosaminidase OS=Salmonella enteritidis PT4 (strain P125109) OX=550537 GN=nagZ PE=3 SV=1 |
| B5FK98 | 6.36e-25 | 75 | 348 | 4 | 277 | Beta-hexosaminidase OS=Salmonella dublin (strain CT_02021853) OX=439851 GN=nagZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
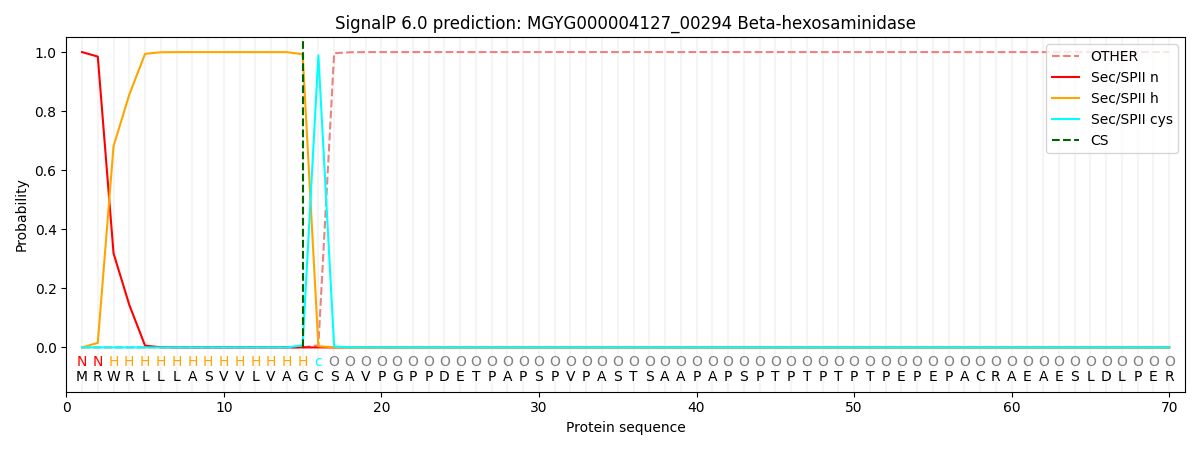
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000069 | 0.000000 | 0.000000 | 0.000000 |
