You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003931_00671
You are here: Home > Sequence: MGYG000003931_00671
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
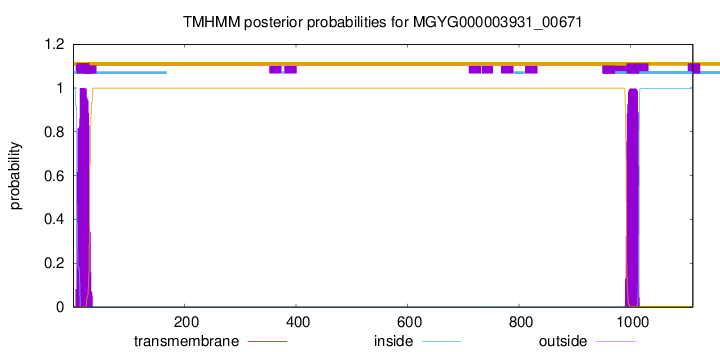
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia_A; Christensenellales; CAG-552; UMGS1880; | |||||||||||
| CAZyme ID | MGYG000003931_00671 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 30677; End: 34015 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 719 | 933 | 9.6e-50 | 0.9722222222222222 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00933 | Glyco_hydro_3 | 1.04e-19 | 721 | 940 | 64 | 290 | Glycosyl hydrolase family 3 N terminal domain. |
| pfam01915 | Glyco_hydro_3_C | 2.35e-15 | 68 | 293 | 1 | 190 | Glycosyl hydrolase family 3 C-terminal domain. This domain is involved in catalysis and may be involved in binding beta-glucan. This domain is found associated with pfam00933. |
| PRK15098 | PRK15098 | 9.75e-11 | 744 | 912 | 117 | 288 | beta-glucosidase BglX. |
| PRK15098 | PRK15098 | 1.56e-10 | 23 | 502 | 345 | 735 | beta-glucosidase BglX. |
| PLN03080 | PLN03080 | 3.94e-10 | 53 | 296 | 388 | 593 | Probable beta-xylosidase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QOS39538.1 | 1.69e-236 | 41 | 1001 | 41 | 999 |
| QUC03329.1 | 1.06e-213 | 59 | 974 | 65 | 939 |
| QOY60704.1 | 1.93e-203 | 18 | 933 | 29 | 908 |
| ADK67194.1 | 4.58e-203 | 58 | 967 | 76 | 970 |
| QOS39539.1 | 1.03e-162 | 52 | 1013 | 52 | 982 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5WUG_A | 1.10e-54 | 66 | 912 | 46 | 748 | Expression,characterization and crystal structure of a novel beta-glucosidase from Paenibacillus barengoltzii [Paenibacillus barengoltzii],5WVP_A Expression, characterization and crystal structure of a novel beta-glucosidase from Paenibacillus barengoltzii [Paenibacillus barengoltzii] |
| 2X40_A | 6.79e-39 | 708 | 933 | 36 | 261 | Structureof beta-glucosidase 3B from Thermotoga neapolitana in complex with glycerol [Thermotoga neapolitana DSM 4359],2X41_A Structure of beta-glucosidase 3B from Thermotoga neapolitana in complex with glucose [Thermotoga neapolitana DSM 4359] |
| 2X42_A | 6.67e-38 | 708 | 933 | 36 | 261 | Structureof beta-glucosidase 3B from Thermotoga neapolitana in complex with alpha-D-glucose [Thermotoga neapolitana DSM 4359] |
| 3AC0_A | 1.99e-28 | 694 | 912 | 9 | 226 | Crystalstructure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus],3AC0_B Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus],3AC0_C Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus],3AC0_D Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus] |
| 5WAB_A | 6.22e-28 | 702 | 912 | 16 | 233 | CrystalStructure of Bifidobacterium adolescentis GH3 beta-glucosidase [Bifidobacterium adolescentis ATCC 15703],5WAB_B Crystal Structure of Bifidobacterium adolescentis GH3 beta-glucosidase [Bifidobacterium adolescentis ATCC 15703],5WAB_C Crystal Structure of Bifidobacterium adolescentis GH3 beta-glucosidase [Bifidobacterium adolescentis ATCC 15703],5WAB_D Crystal Structure of Bifidobacterium adolescentis GH3 beta-glucosidase [Bifidobacterium adolescentis ATCC 15703] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P16084 | 2.59e-64 | 52 | 912 | 24 | 770 | Beta-glucosidase A OS=Butyrivibrio fibrisolvens OX=831 GN=bglA PE=3 SV=1 |
| P15885 | 2.15e-60 | 60 | 937 | 13 | 729 | Beta-glucosidase OS=Ruminococcus albus OX=1264 PE=3 SV=1 |
| Q5BA18 | 1.51e-31 | 694 | 912 | 16 | 233 | Probable beta-glucosidase K OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=bglK PE=2 SV=1 |
| Q5BFG8 | 2.03e-31 | 687 | 917 | 7 | 236 | Beta-glucosidase B OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=bglB PE=1 SV=1 |
| P27034 | 6.97e-30 | 694 | 917 | 5 | 228 | Beta-glucosidase OS=Rhizobium radiobacter OX=358 GN=cbg-1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
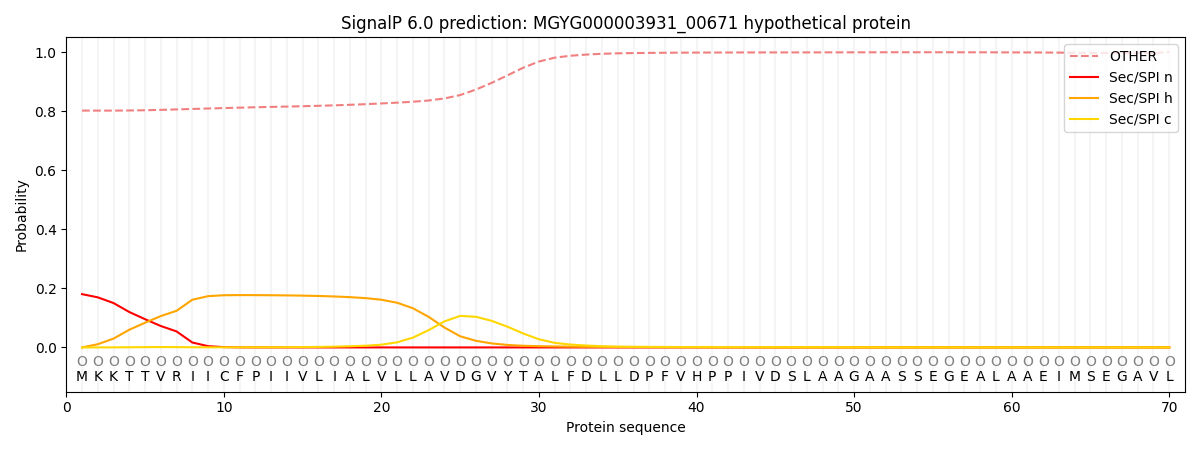
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.811955 | 0.166698 | 0.009500 | 0.000597 | 0.000436 | 0.010822 |