You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003891_01816
You are here: Home > Sequence: MGYG000003891_01816
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
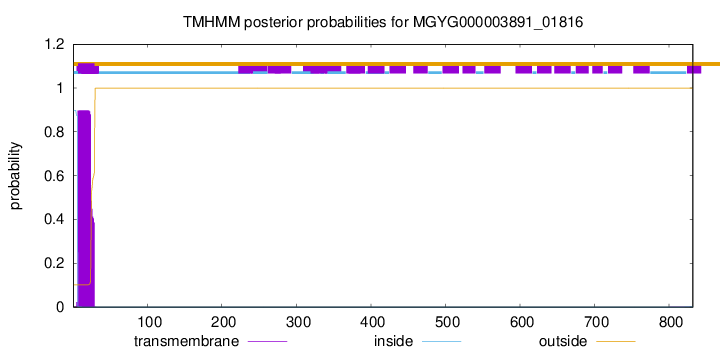
TMHMM annotations
Basic Information help
| Species | UBA11524 sp000437595 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia_A; Christensenellales; CAG-74; UBA11524; UBA11524 sp000437595 | |||||||||||
| CAZyme ID | MGYG000003891_01816 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 4180; End: 6678 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 191 | 393 | 1.1e-18 | 0.9212962962962963 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1472 | BglX | 1.09e-22 | 186 | 545 | 48 | 363 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| PRK15098 | PRK15098 | 2.05e-10 | 97 | 543 | 32 | 417 | beta-glucosidase BglX. |
| pfam00933 | Glyco_hydro_3 | 8.10e-10 | 176 | 462 | 44 | 315 | Glycosyl hydrolase family 3 N terminal domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QTE67443.1 | 5.70e-215 | 65 | 832 | 47 | 838 |
| QTE67446.1 | 2.36e-161 | 53 | 831 | 41 | 793 |
| QOY84902.1 | 4.53e-37 | 72 | 576 | 36 | 528 |
| QTE67433.1 | 3.09e-36 | 48 | 682 | 20 | 644 |
| QRK06309.1 | 1.76e-35 | 59 | 683 | 15 | 632 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5Z9S_A | 2.54e-17 | 98 | 547 | 23 | 413 | Functionaland Structural Characterization of a beta-Glucosidase Involved in Saponin Metabolism from Intestinal Bacteria [Bifidobacterium longum],5Z9S_B Functional and Structural Characterization of a beta-Glucosidase Involved in Saponin Metabolism from Intestinal Bacteria [Bifidobacterium longum] |
| 5M6G_A | 3.06e-16 | 92 | 549 | 40 | 441 | Crystalstructure Glucan 1,4-beta-glucosidase from Saccharopolyspora erythraea [Saccharopolyspora erythraea D] |
| 5YOT_A | 5.07e-16 | 92 | 543 | 7 | 411 | Isoprimeverose-producingenzyme from Aspergillus oryzae in complex with isoprimeverose [Aspergillus oryzae RIB40],5YOT_B Isoprimeverose-producing enzyme from Aspergillus oryzae in complex with isoprimeverose [Aspergillus oryzae RIB40],5YQS_A Isoprimeverose-producing enzyme from Aspergillus oryzae in complex with isoprimeverose [Aspergillus oryzae RIB40],5YQS_B Isoprimeverose-producing enzyme from Aspergillus oryzae in complex with isoprimeverose [Aspergillus oryzae RIB40] |
| 7EAP_A | 5.07e-16 | 92 | 543 | 7 | 411 | ChainA, Fn3_like domain-containing protein [Aspergillus oryzae RIB40] |
| 3WLQ_A | 5.87e-14 | 91 | 549 | 8 | 427 | CrystalStructure Analysis of Plant Exohydrolase [Hordeum vulgare subsp. vulgare],3WLR_A Crystal Structure Analysis of Plant Exohydrolase [Hordeum vulgare subsp. vulgare],6MI1_A CRYSTAL STRUCTURE ANALYSIS OF THE VARIANT PLANT EXOHYDROLASE ARG158ALA-GLU161ALA IN COMPLEX WITH METHYL 6-THIO-BETA-GENTIOBIOSIDE [Hordeum vulgare subsp. vulgare] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q46684 | 1.04e-29 | 55 | 543 | 21 | 502 | Periplasmic beta-glucosidase/beta-xylosidase OS=Dickeya chrysanthemi OX=556 GN=bgxA PE=3 SV=1 |
| B8NGU6 | 2.73e-21 | 91 | 683 | 41 | 609 | Probable beta-glucosidase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=bglC PE=3 SV=1 |
| Q2UFP8 | 8.46e-21 | 86 | 683 | 40 | 613 | Probable beta-glucosidase C OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=bglC PE=3 SV=2 |
| Q5BCC6 | 3.71e-18 | 89 | 683 | 35 | 602 | Beta-glucosidase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=bglC PE=1 SV=1 |
| T2KMH0 | 2.58e-12 | 179 | 533 | 39 | 384 | Beta-xylosidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22130 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
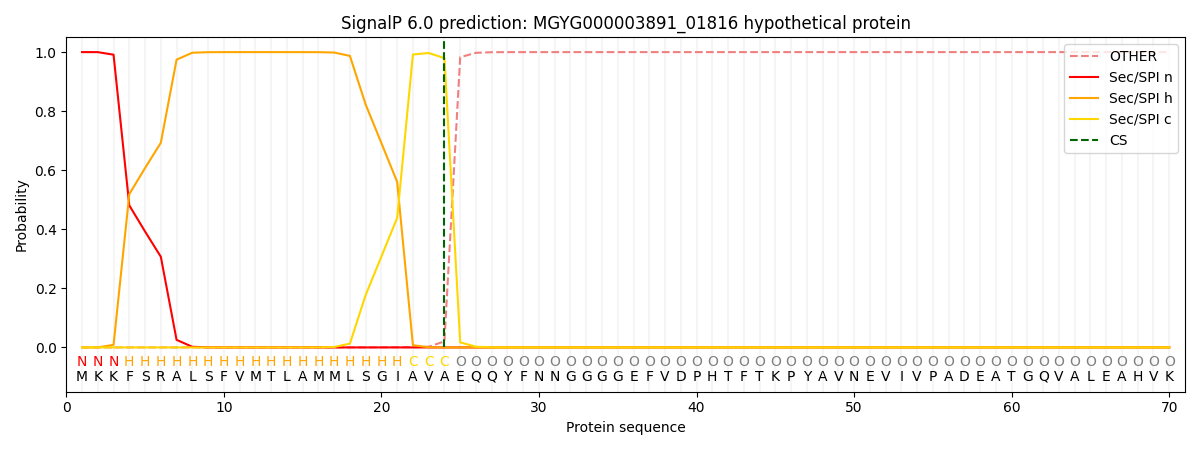
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000279 | 0.998972 | 0.000240 | 0.000173 | 0.000168 | 0.000155 |