You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003891_00753
You are here: Home > Sequence: MGYG000003891_00753
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UBA11524 sp000437595 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia_A; Christensenellales; CAG-74; UBA11524; UBA11524 sp000437595 | |||||||||||
| CAZyme ID | MGYG000003891_00753 | |||||||||||
| CAZy Family | GH106 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 135; End: 3191 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH106 | 35 | 852 | 3.5e-72 | 0.845873786407767 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17132 | Glyco_hydro_106 | 2.21e-21 | 195 | 771 | 343 | 873 | alpha-L-rhamnosidase. |
| cd18241 | BTB_POZ_KLHL11 | 0.003 | 42 | 76 | 63 | 97 | BTB (Broad-Complex, Tramtrack and Bric a brac)/POZ (poxvirus and zinc finger) domain found in Kelch-like protein 11 (KLHL11). KLHL11 is a component of a cullin-RING-based BCR (BTB-CUL3-RBX1) E3 ubiquitin-protein ligase complex that mediates the ubiquitination of target proteins, leading most often to their proteasomal degradation. It contains a BTB domain and kelch repeats, characteristics of a kelch family protein. The BTB/POZ domain is a common protein-protein interaction motif of about 100 amino acids. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGH71057.1 | 8.26e-262 | 29 | 1012 | 80 | 1126 |
| AHF25191.1 | 1.01e-251 | 26 | 1012 | 53 | 1147 |
| QYN20448.1 | 7.06e-130 | 46 | 982 | 63 | 1081 |
| SDG73971.1 | 9.15e-129 | 33 | 982 | 5 | 930 |
| QGH70002.1 | 2.10e-125 | 13 | 1018 | 60 | 1116 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
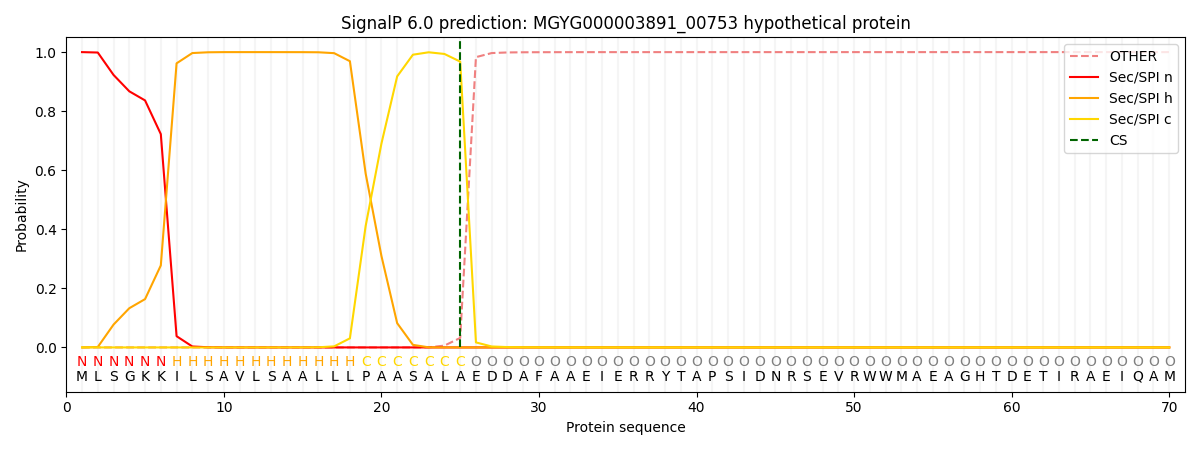
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000346 | 0.998849 | 0.000194 | 0.000213 | 0.000188 | 0.000161 |
