You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003680_01723
You are here: Home > Sequence: MGYG000003680_01723
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp000433175 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp000433175 | |||||||||||
| CAZyme ID | MGYG000003680_01723 | |||||||||||
| CAZy Family | GH43 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 9171; End: 13304 Strand: + | |||||||||||
Full Sequence Download help
| MKKLFLSAII AAATMASSIT SAKDVYVSTS FREPANEGLR FIYSYDGLRW DTIQGSFLRP | 60 |
| EVGTQKVMRD PSIVKGPDGT FHLVWTSSWR GDKGFGYASS KDLIHWSKER LINVMDDPTT | 120 |
| VNVWAPELFY DDVKKQYMIV WASCVPGKFP DYEEDHYNNH RLYYVTTKDF KKFSKAKVLI | 180 |
| DPDFSCIDAT ILKRGDKDYV MVLKDNSRKQ RNLKISFAKS PYGPWSKPSE PFTESFTEGP | 240 |
| TTAKAEGWYY VYYDSYQHGI YGAARTKDFV NFQDRTGAVN FPVGHKHGTV FMASEEIVNN | 300 |
| LIKYNRDAVH YTGSVIATPA RHDGGLSPVV GVHNIQTMRG SAGWLYNHQP MMAYWNNKFY | 360 |
| MHYLTDPRSE HEAPGRTMLQ TSDDGYTWTE PVVLFPEYNV PDGFTKPAWP GVVAKDLKAI | 420 |
| MHQRIGFYVS KSNKLIAMGN YCVAITPKDD PNDGNAIGRV VREIKADGSF GPIYFIYYNH | 480 |
| GFNEKNTDYP YYKRSKDKAF VKACDELLAD PMIRMQWVEE ADRDDEILPL KTPYKAFSGY | 540 |
| TLPDGRKVGL WKHALTSISS DGGNTWRQPV KRAHGFVNSN AKIWGQRLSD GTYATVYNPA | 600 |
| EYRWPLAISL SNDGLEYTTL NLINGEVTPE RHWGLYKSFG PQYTRGILEG NGKPKDGNLW | 660 |
| ITYSNNKEDM WVACVQVPVK LEATEQASGG FGKYKKLADM TDWNIYSPLW APVKLDGEWL | 720 |
| TLSDKDPYDY ARVEKVIPAT KELTVEFDVK AMQNDHGQLN IEFLDAKGNM CSRIVLDSTA | 780 |
| TMKVKGGARY GGMMKHYDVG QAYHIKAVLS VDGHVGTYFV NGKKATARMF DTPVEAITRI | 840 |
| VFRTGNLFDK PDIETPADQT VDMPRADEQD PMATFAIANL TTKSSDTDAN AAVLKYDNYK | 900 |
| HYADRFNTME DENIETYIPN SKSSEWMSKN VPLFDCPDKD MEEIYYFRWW SLRKHIKRTP | 960 |
| VGLGMTEFLV QRSYADKYNL IACAVGHHIM ETRWLRDTTY LHQILNTWYH GNNGKAMTKM | 1020 |
| NKFSSWNPAA VFEAYKVLGD TAFVLGLKPS LEEEYARWKS TNRLANGLYW QGDVQDGMEE | 1080 |
| SISGGRRKQY ARPTINSYMY GNARALAELN RMAGDASKAK EYDLEADTLK SLIQSKLWNE | 1140 |
| KHSFFETVRG DTAAAVREAI GYIPWYFNLP DAKKYDEAWK QLEDEKGFSA PYGLTTAERR | 1200 |
| HPEFRSHGVG KCEWDGAIWP FATSQTLTAF ANYINEYPEP VLGDSTFFKQ MKLYVESQHH | 1260 |
| RGRPYIGEYL DEKNGAWLMG DRERSRYYNH STFADLVITG LVGLRPQADG SIVVNPLVPE | 1320 |
| NTWTYFCLDN VNYRGNVLTI IYDKDGQRYH QGKGFRLLVN GKTVCEREDL GKLTYRK | 1377 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH143 | 304 | 842 | 2.2e-244 | 0.9892086330935251 |
| GH142 | 906 | 1371 | 6.2e-211 | 0.9937369519832986 |
| GH43 | 66 | 291 | 6e-82 | 0.9912663755458515 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd08983 | GH43_Bt3655-like | 4.38e-74 | 56 | 304 | 7 | 261 | Glycosyl hydrolase family 43 protein such as Bacteroides thetaiotaomicron VPI-5482 arabinofuranosidase Bt3655. This glycosyl hydrolase family 43 (GH43)-like family includes the characterized arabinofuranosidases (EC 3.2.1.55): Bacteroides thetaiotaomicron VPI-5482 (Bt3655;BT_3655) and Penicillium chrysogenum 31B Abf43B, as well as Bifidobacterium adolescentis ATCC 15703 beta-xylosidase (EC 3.2.1.37) BAD_1527. It belongs to the glycosyl hydrolase clan F (according to carbohydrate-active enzymes database (CAZY)) which includes family 43 (GH43) and 62 (GH62) families. GH43 includes enzymes with beta-xylosidase (EC 3.2.1.37), beta-1,3-xylosidase (EC 3.2.1.-), alpha-L-arabinofuranosidase (EC 3.2.1.55), arabinanase (EC 3.2.1.99), xylanase (EC 3.2.1.8), endo-alpha-L-arabinanases (beta-xylanases) and galactan 1,3-beta-galactosidase (EC 3.2.1.145) activities. GH43 are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. Many GH43 enzymes display both alpha-L-arabinofuranosidase and beta-D-xylosidase activity using aryl-glycosides as substrates. A common structural feature of GH43 enzymes is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| cd08978 | GH_F | 6.00e-13 | 69 | 256 | 1 | 191 | Glycosyl hydrolase families 43 and 62 form CAZY clan GH-F. This glycosyl hydrolase clan F (according to carbohydrate-active enzymes database (CAZY)) includes family 43 (GH43) and 62 (GH62). GH43 includes enzymes with beta-xylosidase (EC 3.2.1.37), beta-1,3-xylosidase (EC 3.2.1.-), alpha-L-arabinofuranosidase (EC 3.2.1.55), arabinanase (EC 3.2.1.99), xylanase (EC 3.2.1.8), endo-alpha-L-arabinanases (beta-xylanases) and galactan 1,3-beta-galactosidase (EC 3.2.1.145) activities. GH62 includes enzymes characterized as arabinofuranosidases (alpha-L-arabinofuranosidases; EC 3.2.1.55) that specifically cleave either alpha-1,2 or alpha-1,3-L-arabinofuranose side chains from xylans. GH43 are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. Many of the enzymes in this family display both alpha-L-arabinofuranosidase and beta-D-xylosidase activity using aryl-glycosides as substrates. GH62 are also predicted to be inverting enzymes. A common structural feature of both, GH43 and GH62 enzymes, is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| cd08986 | GH43-like | 3.43e-09 | 68 | 227 | 2 | 218 | Glycosyl hydrolase family 43 protein; uncharacterized. This glycosyl hydrolase family 43 (GH43)-like subfamily includes uncharacterized enzymes similar to those with beta-1,4-xylosidase (xylan 1,4-beta-xylosidase; EC 3.2.1.37), beta-1,3-xylosidase (EC 3.2.1.-), alpha-L-arabinofuranosidase (EC 3.2.1.55), arabinanase (EC 3.2.1.99), xylanase (EC 3.2.1.8), endo-alpha-L-arabinanase and galactan 1,3-beta-galactosidase (EC 3.2.1.145) activities. These are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. Many of the enzymes in this family display both alpha-L-arabinofuranosidase and beta-D-xylosidase activity using aryl-glycosides as substrates. A common structural feature of GH43 enzymes is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| pfam13088 | BNR_2 | 1.30e-07 | 540 | 614 | 168 | 247 | BNR repeat-like domain. This family of proteins contains BNR-like repeats suggesting these proteins may act as sialidases. |
| pfam04616 | Glyco_hydro_43 | 1.11e-06 | 70 | 253 | 12 | 191 | Glycosyl hydrolases family 43. The glycosyl hydrolase family 43 contains members that are arabinanases. Arabinanases hydrolyze the alpha-1,5-linked L-arabinofuranoside backbone of plant cell wall arabinans. The structure of arabinanase Arb43A from Cellvibrio japonicus reveals a five-bladed beta-propeller fold. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT77852.1 | 0.0 | 306 | 1373 | 21 | 1100 |
| ALJ45260.1 | 0.0 | 304 | 1373 | 21 | 1099 |
| QRQ56889.1 | 0.0 | 304 | 1373 | 21 | 1099 |
| SCV09596.1 | 0.0 | 304 | 1373 | 21 | 1099 |
| QRN01961.1 | 0.0 | 304 | 1373 | 21 | 1099 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5MQR_A | 0.0 | 306 | 1373 | 25 | 1101 | SialidaseBT_1020 [Bacteroides thetaiotaomicron] |
| 5MQS_A | 0.0 | 306 | 1373 | 25 | 1101 | SialidaseBT_1020 [Bacteroides thetaiotaomicron] |
| 6M5A_A | 1.14e-24 | 924 | 1375 | 303 | 811 | Crystalstructure of GH121 beta-L-arabinobiosidase HypBA2 from Bifidobacterium longum [Bifidobacterium longum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| E8MGH9 | 1.08e-23 | 924 | 1375 | 334 | 842 | Beta-L-arabinobiosidase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=hypBA2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
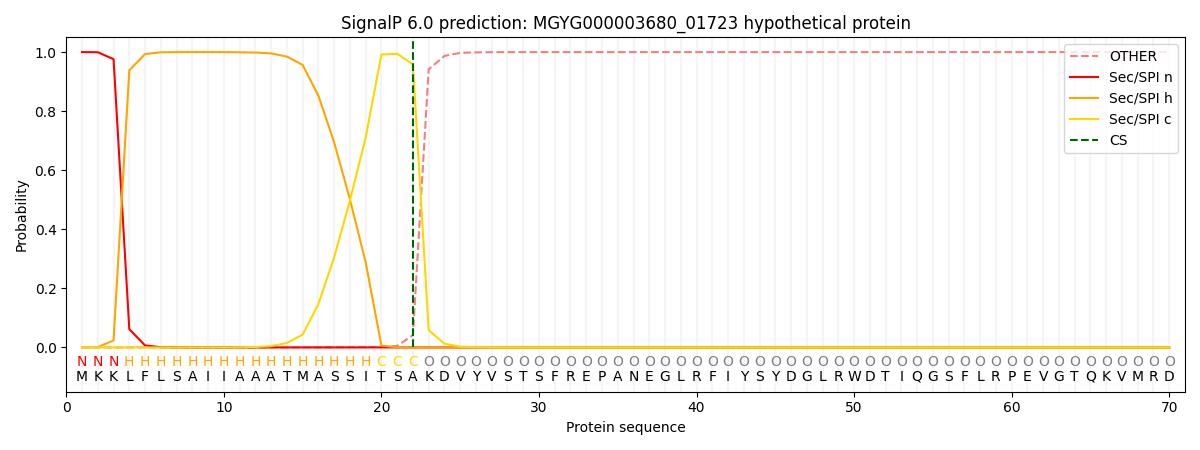
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000367 | 0.998897 | 0.000199 | 0.000202 | 0.000171 | 0.000151 |
