You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003639_00209
You are here: Home > Sequence: MGYG000003639_00209
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
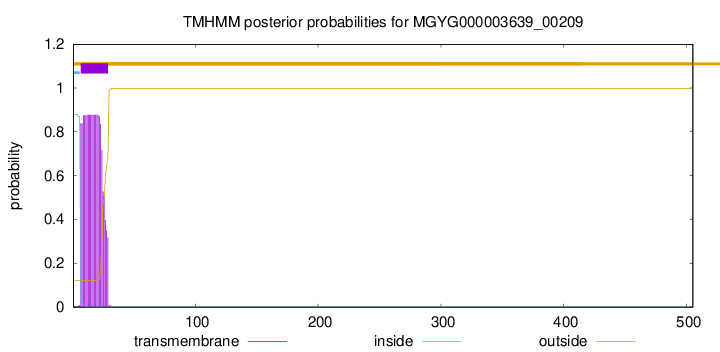
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Kiritimatiellae; RFP12; UBA1067; UBA1067; | |||||||||||
| CAZyme ID | MGYG000003639_00209 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 694; End: 2211 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 3.06e-21 | 193 | 466 | 11 | 271 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 1.63e-16 | 199 | 391 | 71 | 271 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM46198.1 | 9.79e-138 | 1 | 504 | 1 | 499 |
| AVM45719.1 | 1.63e-135 | 25 | 504 | 65 | 549 |
| AVM43579.1 | 4.92e-134 | 30 | 504 | 50 | 526 |
| AVM46390.1 | 1.71e-128 | 80 | 504 | 76 | 501 |
| AVM45720.1 | 5.16e-128 | 30 | 502 | 84 | 559 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3AMC_A | 4.50e-15 | 253 | 468 | 79 | 288 | Crystalstructures of Thermotoga maritima Cel5A, apo form and dimer/au [Thermotoga maritima MSB8],3AMC_B Crystal structures of Thermotoga maritima Cel5A, apo form and dimer/au [Thermotoga maritima MSB8],3AMD_A Crystal structures of Thermotoga maritima Cel5A, apo form and tetramer/au [Thermotoga maritima MSB8],3AMD_B Crystal structures of Thermotoga maritima Cel5A, apo form and tetramer/au [Thermotoga maritima MSB8],3AMD_C Crystal structures of Thermotoga maritima Cel5A, apo form and tetramer/au [Thermotoga maritima MSB8],3AMD_D Crystal structures of Thermotoga maritima Cel5A, apo form and tetramer/au [Thermotoga maritima MSB8],3MMU_A Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_B Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_C Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_D Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_E Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_F Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_G Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_H Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMW_A Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMW_B Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMW_C Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMW_D Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima] |
| 7EC9_A | 2.48e-14 | 204 | 438 | 39 | 274 | ChainA, Endoglucanase [Thermotoga maritima MSB8],7EC9_B Chain B, Endoglucanase [Thermotoga maritima MSB8],7EFZ_A Chain A, Endoglucanase [Thermotoga maritima MSB8],7EFZ_B Chain B, Endoglucanase [Thermotoga maritima MSB8] |
| 3AMG_A | 2.65e-14 | 253 | 468 | 79 | 288 | Crystalstructures of Thermotoga maritima Cel5A in complex with Cellobiose substrate, mutant form [Thermotoga maritima MSB8],3AMG_B Crystal structures of Thermotoga maritima Cel5A in complex with Cellobiose substrate, mutant form [Thermotoga maritima MSB8] |
| 3W0K_A | 3.01e-14 | 210 | 486 | 31 | 309 | CrystalStructure of a glycoside hydrolase [Caldanaerobius polysaccharolyticus],3W0K_B Crystal Structure of a glycoside hydrolase [Caldanaerobius polysaccharolyticus] |
| 3NCO_A | 1.19e-13 | 203 | 468 | 42 | 295 | Crystalstructure of FnCel5A from F. nodosum Rt17-B1 [Fervidobacterium nodosum Rt17-B1],3NCO_B Crystal structure of FnCel5A from F. nodosum Rt17-B1 [Fervidobacterium nodosum Rt17-B1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P16169 | 1.67e-18 | 204 | 471 | 33 | 296 | Cellodextrinase A OS=Ruminococcus flavefaciens OX=1265 GN=celA PE=3 SV=3 |
| B0XN12 | 3.49e-08 | 204 | 381 | 98 | 271 | Probable glucan 1,3-beta-glucosidase A OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=exgA PE=3 SV=1 |
| Q4WK60 | 1.08e-07 | 204 | 381 | 98 | 271 | Probable glucan 1,3-beta-glucosidase A OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=exgA PE=3 SV=1 |
| P54583 | 4.22e-07 | 246 | 471 | 135 | 362 | Endoglucanase E1 OS=Acidothermus cellulolyticus (strain ATCC 43068 / DSM 8971 / 11B) OX=351607 GN=Acel_0614 PE=1 SV=1 |
| A1D4Q5 | 1.03e-06 | 241 | 381 | 129 | 271 | Probable glucan 1,3-beta-glucosidase A OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=exgA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001526 | 0.997418 | 0.000361 | 0.000219 | 0.000213 | 0.000232 |