You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003556_01477
You are here: Home > Sequence: MGYG000003556_01477
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
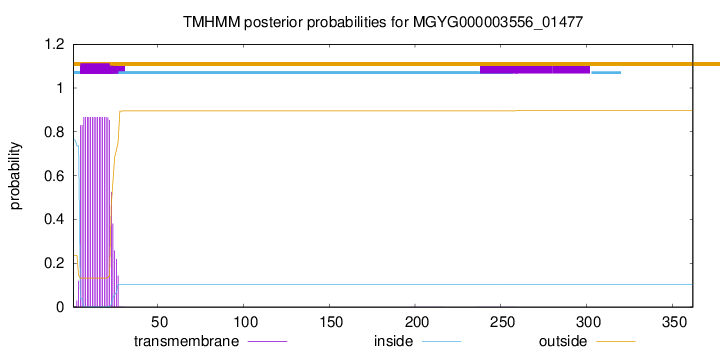
TMHMM annotations
Basic Information help
| Species | Alistipes sp900769745 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; Alistipes sp900769745 | |||||||||||
| CAZyme ID | MGYG000003556_01477 | |||||||||||
| CAZy Family | CE15 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1862; End: 2950 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE15 | 56 | 362 | 2.9e-24 | 0.9739776951672863 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0412 | DLH | 3.99e-05 | 118 | 251 | 8 | 140 | Dienelactone hydrolase [Secondary metabolites biosynthesis, transport and catabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDH54042.1 | 7.80e-147 | 23 | 361 | 28 | 383 |
| QMW85982.1 | 3.15e-146 | 31 | 361 | 36 | 383 |
| QQA08270.1 | 3.15e-146 | 31 | 361 | 36 | 383 |
| ALJ44193.1 | 3.15e-146 | 31 | 361 | 36 | 383 |
| AAO78713.1 | 3.15e-146 | 31 | 361 | 36 | 383 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7NN3_A | 1.26e-12 | 58 | 341 | 30 | 345 | ChainA, Beta-xylanase [Caldicellulosiruptor kristjanssonii I77R1B],7NN3_B Chain B, Beta-xylanase [Caldicellulosiruptor kristjanssonii I77R1B],7NN3_C Chain C, Beta-xylanase [Caldicellulosiruptor kristjanssonii I77R1B],7NN3_D Chain D, Beta-xylanase [Caldicellulosiruptor kristjanssonii I77R1B] |
| 6HSW_A | 1.90e-08 | 60 | 312 | 74 | 369 | ACE15 glucuronoyl esterase from Teredinibacter turnerae T7901 [Teredinibacter turnerae T7901],6HSW_B A CE15 glucuronoyl esterase from Teredinibacter turnerae T7901 [Teredinibacter turnerae T7901],6HSW_C A CE15 glucuronoyl esterase from Teredinibacter turnerae T7901 [Teredinibacter turnerae T7901] |
| 6GRW_A | 3.08e-07 | 200 | 312 | 207 | 328 | GlucuronoylEsterase from Opitutus terrae (Au derivative) [Opitutus terrae PB90-1],6GS0_A Native Glucuronoyl Esterase from Opitutus terrae [Opitutus terrae PB90-1] |
| 6SYU_A | 3.18e-07 | 200 | 312 | 225 | 346 | Thewild type glucuronoyl esterase OtCE15A from Opitutus terrae in complex with xylobiose [Opitutus terrae],6T0I_A The wild type glucuronoyl esterase OtCE15A from Opitutus terrae in complex with the aldotetrauronic acid XUX [Opitutus terrae] |
| 6SZ0_A | 3.18e-07 | 200 | 312 | 225 | 346 | Theglucuronoyl esterase OtCE15A H408A variant from Opitutus terrae [Opitutus terrae PB90-1],6SZ4_A The glucuronoyl esterase OtCE15A H408A variant from Opitutus terrae in complex with, and covalently linked to, D-glucuronate [Opitutus terrae PB90-1] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
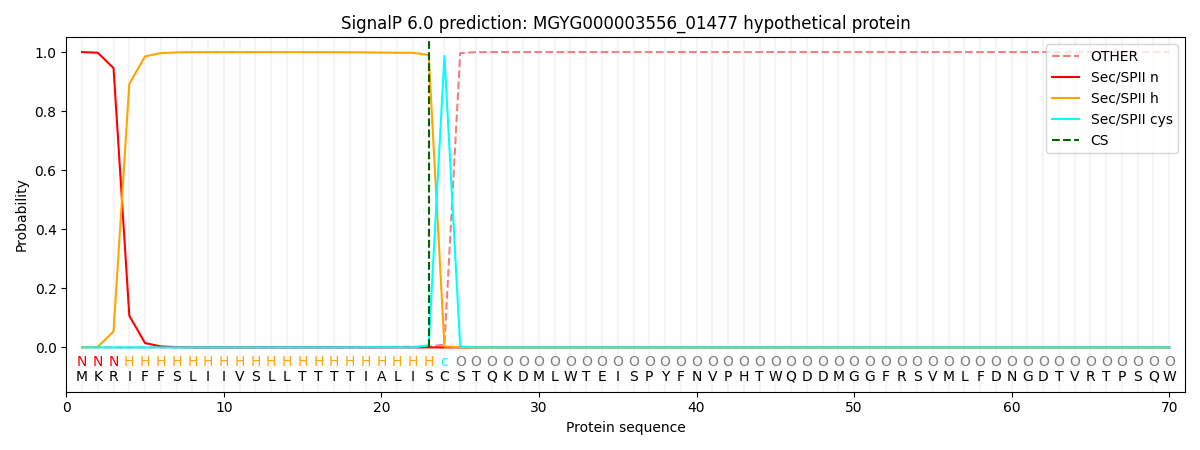
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000015 | 1.000047 | 0.000000 | 0.000000 | 0.000000 |