You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003528_02564
You are here: Home > Sequence: MGYG000003528_02564
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAAGGB01 sp900769285 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; UBA1407; UBA1407; CAAGGB01; CAAGGB01 sp900769285 | |||||||||||
| CAZyme ID | MGYG000003528_02564 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1055; End: 4237 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 396 | 861 | 3.1e-34 | 0.5159574468085106 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 1.96e-17 | 499 | 891 | 116 | 497 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| pfam00703 | Glyco_hydro_2 | 1.48e-08 | 555 | 665 | 1 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| pfam02836 | Glyco_hydro_2_C | 2.24e-07 | 674 | 880 | 8 | 202 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| PRK10150 | PRK10150 | 0.001 | 560 | 797 | 176 | 417 | beta-D-glucuronidase; Provisional |
| COG1842 | PspA | 0.006 | 320 | 366 | 87 | 132 | Phage shock protein A [Transcription, Signal transduction mechanisms]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM44993.1 | 6.10e-227 | 11 | 932 | 54 | 1038 |
| QUH31336.1 | 1.04e-17 | 399 | 888 | 2 | 463 |
| QQT35320.1 | 2.71e-15 | 392 | 893 | 32 | 498 |
| VIP08421.1 | 1.19e-14 | 24 | 223 | 590 | 766 |
| AIQ71089.1 | 1.62e-14 | 542 | 929 | 176 | 555 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5T98_A | 1.52e-09 | 472 | 802 | 88 | 422 | Crystalstructure of BuGH2Awt [Bacteroides uniformis],5T98_B Crystal structure of BuGH2Awt [Bacteroides uniformis],5T99_A Crystal structure of BuGH2Awt in complex with Galactoisofagomine [Bacteroides uniformis],5T99_B Crystal structure of BuGH2Awt in complex with Galactoisofagomine [Bacteroides uniformis] |
| 7CWD_A | 7.79e-09 | 459 | 857 | 68 | 451 | ChainA, beta-glalactosidase [Niallia circulans],7CWI_A Chain A, beta-galactosidase [Niallia circulans] |
| 4CU6_A | 3.58e-07 | 627 | 798 | 253 | 429 | Unravellingthe multiple functions of the architecturally intricate Streptococcus pneumoniae beta-galactosidase, BgaA [Streptococcus pneumoniae TIGR4],4CU7_A Unravelling the multiple functions of the architecturally intricate Streptococcus pneumoniae beta-galactosidase, BgaA [Streptococcus pneumoniae TIGR4],4CU8_A Unravelling the multiple functions of the architecturally intricate Streptococcus pneumoniae beta-galactosidase, BgaA [Streptococcus pneumoniae TIGR4] |
| 4CUC_A | 3.58e-07 | 627 | 798 | 253 | 429 | Unravellingthe multiple functions of the architecturally intricate Streptococcus pneumoniae beta-galactosidase, BgaA. [Streptococcus pneumoniae TIGR4] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KM09 | 5.37e-07 | 638 | 797 | 278 | 432 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22050 PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
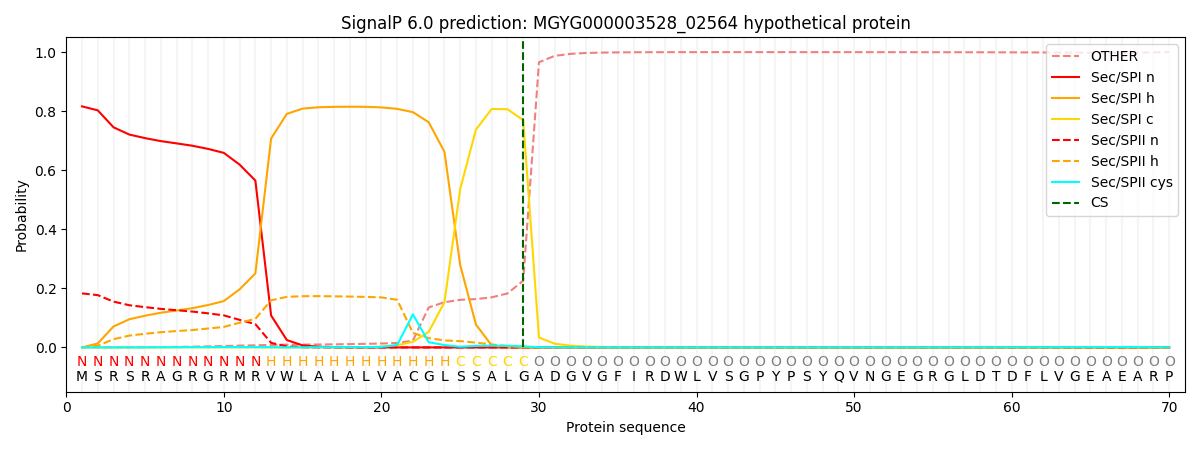
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001786 | 0.805693 | 0.191496 | 0.000397 | 0.000318 | 0.000293 |

