You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003515_00110
Basic Information
help
| Species |
Prevotella sp900769275
|
| Lineage |
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900769275
|
| CAZyme ID |
MGYG000003515_00110
|
| CAZy Family |
GH143 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000003515 |
2052370 |
MAG |
Fiji |
Oceania |
|
| Gene Location |
Start: 26298;
End: 29606
Strand: -
|
No EC number prediction in MGYG000003515_00110.
| Family |
Start |
End |
Evalue |
family coverage |
| GH143 |
25 |
575 |
8.8e-224 |
0.9838129496402878 |
| GH142 |
635 |
1100 |
4.7e-180 |
0.9812108559498957 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| pfam13088
|
BNR_2 |
7.41e-08 |
274 |
351 |
166 |
247 |
BNR repeat-like domain. This family of proteins contains BNR-like repeats suggesting these proteins may act as sialidases. |
| COG3408
|
GDB1 |
0.002 |
744 |
1059 |
330 |
625 |
Glycogen debranching enzyme (alpha-1,6-glucosidase) [Carbohydrate transport and metabolism]. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 5MQR_A
|
0.0 |
20 |
1098 |
19 |
1090 |
SialidaseBT_1020 [Bacteroides thetaiotaomicron] |
| 5MQS_A
|
0.0 |
20 |
1098 |
19 |
1090 |
SialidaseBT_1020 [Bacteroides thetaiotaomicron] |
This protein is predicted as SP
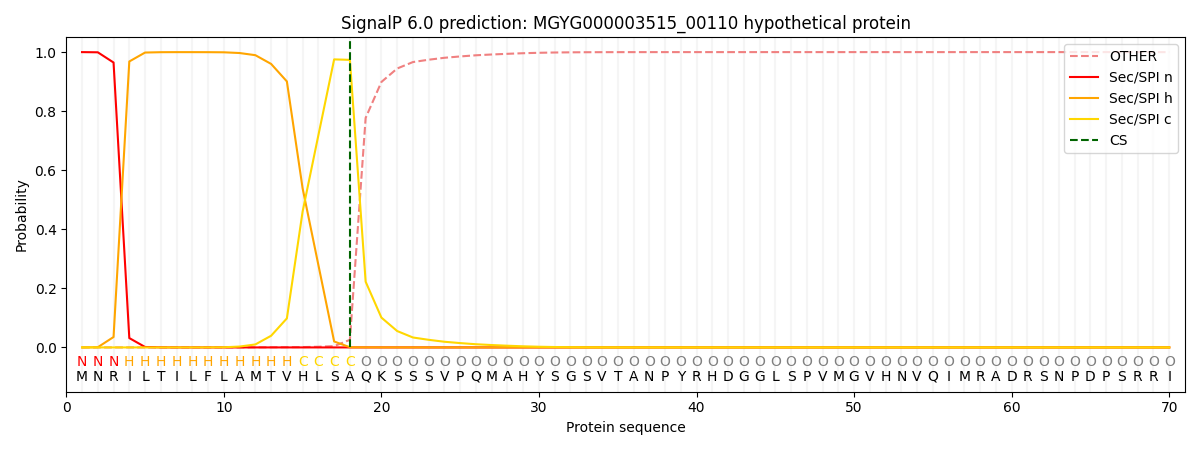
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.000425
|
0.998659
|
0.000413
|
0.000173
|
0.000160
|
0.000151
|
There is no transmembrane helices in MGYG000003515_00110.

